Reversine Although SGs are referred to as stalled translation preinitiation complexes, as well as on intensity and type of the stress. For example, SGs induced by a high concentration of ethanol in S. cerevisiae contain only the eIF3c/Nip1 subunit of the eIF3 complex and SGs induced by a prolonged glucose deprivation do not harbor the eIF3 complex at all. With respect to the intensity of the stress, treatment with a low concentration of NaN3 does not affect the distribution of eIF3a but at higher concentration this drug induces eIF3a accumulation in SGs. 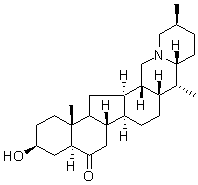 We show here that SGs induced by the robust heat shock in S. cerevisiae contain translation elongation factors eEF3 and eEF1Bc2 together with translation termination factors eRF1 and eRF3. These factors have never been observed in SGs of any other eukaryotic cell. However, the termination factors have been found to accumulate in P-bodies. Those authors have concluded that presence of translation termination factors in Pbodies is BAY-60-7550 clinical trial coupled to the P-bodies assembly. A similar role could be suggested for presence of these factors in heat-induced SGs. The proteins with self-aggregation domain, like TIA-1 or TIAR in mammalian cells, have been described to influence dynamics of SGs. A newly identified component of the heat-induced SGs in S.cerevisiae, Sup35p, possesses a prion-like domain at the N-terminus. Sup35p can thus convert into the prion form, known as. The N-terminal part of the protein is indispensable for the prion formation and maintenance. Similarly to a situation in mammalian cells we found that rather a non-prion part of Sup35p is responsible for accumulation of the protein in SGs. However, observations that SGs are formed even in the absence of the N-terminal prion-like domain of Sup35p indicate that unlike in mammals, the assembly of heatinduced SGs in S. cerevisiae is not driven by these “prion” structural elements. This hypothesis is also supported by our earlier findings that heat-induced SGs are formed even in the absence of yeast orthologs of mammalian TIA-1 and TIAR proteins, Ngr1 and Pub1 proteins in S. cerevisiae. Translation termination factors eRF1 and eRF3 are responsible for effective termination of translation. In addition, they seem to be required for an effective function of the fungal-specific elongation factor eEF3 in recycling of the translation posttermination complexes after the release of newly synthetized peptide chains. In this respect, identification of elongation and termination factors in heatinduced SGs may indicate that these SGs are composed of translation posttermination complexes stalled before the ribosome recycling step. However, we did not observe any accumulation of several essential proteins of the 60S ribosomal subunits under robust heat shock and Grousl et al.. In addition, all the published information on recycling of the translation posttermination complexes comes from in vitro experiments only. Therefore, it is currently unclear, how recycling is catalyzed in vivo and the reasons for presence of the translation elongation and the termination factors in robust heat shockinduced SGs remain to be elucidated. Whereas different roles for SGs and P-bodies in cell survival upon heat stress conditions could be suggested, both accumulations are always closely spatially and functionally intertwined. In S. cerevisiae cells, P-bodies promote formation of SGs.
We show here that SGs induced by the robust heat shock in S. cerevisiae contain translation elongation factors eEF3 and eEF1Bc2 together with translation termination factors eRF1 and eRF3. These factors have never been observed in SGs of any other eukaryotic cell. However, the termination factors have been found to accumulate in P-bodies. Those authors have concluded that presence of translation termination factors in Pbodies is BAY-60-7550 clinical trial coupled to the P-bodies assembly. A similar role could be suggested for presence of these factors in heat-induced SGs. The proteins with self-aggregation domain, like TIA-1 or TIAR in mammalian cells, have been described to influence dynamics of SGs. A newly identified component of the heat-induced SGs in S.cerevisiae, Sup35p, possesses a prion-like domain at the N-terminus. Sup35p can thus convert into the prion form, known as. The N-terminal part of the protein is indispensable for the prion formation and maintenance. Similarly to a situation in mammalian cells we found that rather a non-prion part of Sup35p is responsible for accumulation of the protein in SGs. However, observations that SGs are formed even in the absence of the N-terminal prion-like domain of Sup35p indicate that unlike in mammals, the assembly of heatinduced SGs in S. cerevisiae is not driven by these “prion” structural elements. This hypothesis is also supported by our earlier findings that heat-induced SGs are formed even in the absence of yeast orthologs of mammalian TIA-1 and TIAR proteins, Ngr1 and Pub1 proteins in S. cerevisiae. Translation termination factors eRF1 and eRF3 are responsible for effective termination of translation. In addition, they seem to be required for an effective function of the fungal-specific elongation factor eEF3 in recycling of the translation posttermination complexes after the release of newly synthetized peptide chains. In this respect, identification of elongation and termination factors in heatinduced SGs may indicate that these SGs are composed of translation posttermination complexes stalled before the ribosome recycling step. However, we did not observe any accumulation of several essential proteins of the 60S ribosomal subunits under robust heat shock and Grousl et al.. In addition, all the published information on recycling of the translation posttermination complexes comes from in vitro experiments only. Therefore, it is currently unclear, how recycling is catalyzed in vivo and the reasons for presence of the translation elongation and the termination factors in robust heat shockinduced SGs remain to be elucidated. Whereas different roles for SGs and P-bodies in cell survival upon heat stress conditions could be suggested, both accumulations are always closely spatially and functionally intertwined. In S. cerevisiae cells, P-bodies promote formation of SGs.
Enrollments of patients with different infection spanning over more than a single influenza season
A disadvantage is that while the global test identifies biomarker groupings that are significant, it does not provide information on which PF-2341066 c-Met inhibitor markers are driving the statistical significance. Also, it is not an optimal test procedure if the association of some markers with disease outcome is positive and that of some other markers is negative. Thus, we also carried out a likelihood ratio test that tested the significance of adding all the markers in a functional category to a base model that only included age, duration of symptoms and geographic region. In an influenza challenge study reported in 1998 in which normal human volunteers were experimentally infected with a seasonal influenza A virus, Hayden et al. found that both IL-6 and IFN-alpha levels in nasal lavage fluids peaked early in the course of infection and correlated directly with viral titers, temperature, mucus production, and symptom Nilotinib scores. Several other clinical studies in adults or children have been reported by investigators in which the addition of various serum or nasal biomarker level measurements to the standard clinical evaluation appeared to add to the diagnostic certainty of respiratory virus infection with seasonal influenza. In a number of generally cross-sectional studies focusing either partially or exclusively upon confirmed cases of Apdm09 virus infection in specific geographic areas, potential correlations between various cytokine levels and disease severity have been reported. Investigators from Mexico reported that Apdm09 virus infection resulted in stronger in vitro upregulation of IL-6, CCL3, and CXCL8 in 72-hour cell cultures as well as elevated serum levels of 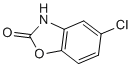 IL-6, CXCL8, and certain other cytokines in individuals infected with this subtype compared to those with seasonal influenza virus infection. In separate publications a group in Hong Kong reported that elevated levels of IL-6, CXCL8, CCL2, and sTNFR-1 correlated with severe cases of Apdm09 virus infection overall and, in particular, with the extent and severity of influenza-associated pneumonia. Similar findings concerning elevated IP-10 and IL-6 levels in cases of pediatric pneumonia were reported from Korea. At least two groups from mainland China have described similar relationships for several of the pro-inflammatory cytokines withApdm09 virus infection, and separate groups from Spain, Italy, and Romania have also described correlations between elevated levels of various cytokines such as IL-6, IL-15, and TNF-a and the severity of disease outcomes in patients with confirmed Apdm09 virus infection. Most recently, a group from Canada has found IL-6 to be an important feature of the host response in both humans and mice infected with Apdm09 virus and, in the former case, found elevated IL-6 levels to be an important predictor of severe disease. Biomarker analyses from our two large ongoing international studies of influenza described here have strengthened and extended these observations in important ways. A major virtue of the present studies is that these data were collected prospectively according to a common data-set and with defined periods of follow-up to assess disease progression, samples have been garnered from a relatively large number of patients living in geographically disparate regions of the world and analyzed through common central laboratory systems using standardized methodologies.
IL-6, CXCL8, and certain other cytokines in individuals infected with this subtype compared to those with seasonal influenza virus infection. In separate publications a group in Hong Kong reported that elevated levels of IL-6, CXCL8, CCL2, and sTNFR-1 correlated with severe cases of Apdm09 virus infection overall and, in particular, with the extent and severity of influenza-associated pneumonia. Similar findings concerning elevated IP-10 and IL-6 levels in cases of pediatric pneumonia were reported from Korea. At least two groups from mainland China have described similar relationships for several of the pro-inflammatory cytokines withApdm09 virus infection, and separate groups from Spain, Italy, and Romania have also described correlations between elevated levels of various cytokines such as IL-6, IL-15, and TNF-a and the severity of disease outcomes in patients with confirmed Apdm09 virus infection. Most recently, a group from Canada has found IL-6 to be an important feature of the host response in both humans and mice infected with Apdm09 virus and, in the former case, found elevated IL-6 levels to be an important predictor of severe disease. Biomarker analyses from our two large ongoing international studies of influenza described here have strengthened and extended these observations in important ways. A major virtue of the present studies is that these data were collected prospectively according to a common data-set and with defined periods of follow-up to assess disease progression, samples have been garnered from a relatively large number of patients living in geographically disparate regions of the world and analyzed through common central laboratory systems using standardized methodologies.
Typical examples of yeast SGs are those involved in organ dysfunctions
TLR5 was up-regulated only after chemotherapy. This suggests that innate immune responses may be somewhat different between early and advanced stage NSCLC patients. These three genes may be candidate biomarkers to predict tumor progression and response to chemotherapy. SLC22A4, which is an organic cation transporter and has been previously identified to confer cellular uptake and sensitivity to anti-tumor drugs, may play a role in the pharmacokinetics of CDDP and GEM. The Syn2 gene has been implicated in synaptogenesis, neurotransmitter release, and the localization of nitric oxide synthase. The relationship between Syn2 and Orbifloxacin cancer has not yet been reported and needs to be clarified in future studies. The present study is biased and has several limitations. First, we used PBMC rather than immune cells in cancer tissues to represent the tumor-induced alteration of the immune system. Genetic Lomitapide Mesylate signatures from PBMC may be dynamically influenced by blood-based immune effector cells, primary tumor, metastatic tumor, and rare tumor cells occasionally detected in blood from cancer patients. However, gene expression profiles derived from PBMC have been suggested as a promising tool for the early detection or prediction of prognosis in cancer patients in several previous studies, implying that a similar regulation of genes may be present in immune cells of cancer tissues and peripheral blood from cancer patients. Reactivity to environmental changes is a key property of all living organisms that enables them to survive and develop. Cells undergo adaptation to stress conditions via various mechanisms. To save energy, general translation is reduced and the expression of stress response-specific genes is triggered. New ribonucleoprotein complexes are formed, through which the fate of mRNA molecules and translation machinery components is regulated. Major accumulations of cytoplasmic RNP complexes that have been recognized in higher eukaryotes – processing bodies and stress granules have also been found in yeasts. Whilst P-bodies are present even in unstressed cells and they become dominant upon stress, SGs are formed only in stressed cells. Besides translationally repressed mRNA molecules, P-bodies also contain proteins involved in mRNA degradation, translation repression, mRNA quality control and other functions. However, nowadays it is evident that the function of P-bodies is far more complex than merely mRNA degradation and is still not well understood. The composition of SGs is influenced by the particular stress conditions and depends on the organism being subjected to the stress. It is generally accepted that major SGs components are stalled translation preinitiation complexes, which contain molecules of mRNA, small ribosomal subunits and several translation initiation factors. Besides these, they may contain a few other factors, which could be found either in SGs or P-bodies. Thus, P-bodies and SGs represent two distinct types of RNP assemblies, which can share some components and 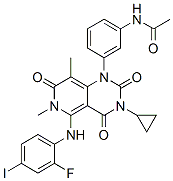 can be in spatial contact, but differ in their role in cell adaptation to environmental changes, which is realized via translation regulation and mRNA metabolism. Analogous to mammalian cells, it is thought that yeast SGs take part in regulation of translation, sorting and storage of mRNA molecules, and the preservation of selected translation factors and mRNA molecules against an influence of a stress. Recently, they have been shown to control TORC1 signaling.
can be in spatial contact, but differ in their role in cell adaptation to environmental changes, which is realized via translation regulation and mRNA metabolism. Analogous to mammalian cells, it is thought that yeast SGs take part in regulation of translation, sorting and storage of mRNA molecules, and the preservation of selected translation factors and mRNA molecules against an influence of a stress. Recently, they have been shown to control TORC1 signaling.
They control a vast array of physiological CTX is well known as an antibacterial drug but less as an antimalarial
Nevertheless, considering the reports on the impact of CTX on malaria, both in HIV-infected and uninfected individuals, we reviewed the available evidence on safety and efficacy of CTX as an antimalarial for both preventive and curative use. It has been well established that exposure to heavy metals including a number of environmental pollutants can cause cellular damages through the formation of highly reactive substances such as reactive oxygen species. ROS show a wide range of pathophysiology. The present study established that exposure to lead nitrate Pb significantly increased ROS formation, enhanced oxidative stress and induced apoptosis in the liver tissue of experimental mice. This adverse effect of Pb, however, could be eliminated by mangiferin treatment probably because of its strong free radical scavenging activity. Besides dietary antioxidants, the body depends on several endogenous defense mechanisms to protect against ROS-induced cell damage. Among these antioxidant molecules, SOD and CAT jointly play important roles in the Orbifloxacin exclusion of ROS. With the purpose of removing excess free radicals from the system, GST and GPx use GSH in their course of reactions. Diminish in GSH content because of oxidative stress reduce the actions of GST and GPx with a concomitant 4-(Benzyloxy)phenol decrease in the activity of GSH stimulating enzyme, GR. The sulfhydryl group of GSH directly binds to heavy metals due to a high affinity for sulfhydryl groups. Lead, arsenic and mercury effectively inactivate the glutathione molecule so it is unavailable as an antioxidant or as a substrate in liver metabolism. In the present study, we found that Pb exposure decreased the activities of the antioxidant enzymes, CAT, SOD, GST, GPX and GR in addition to the level of GSH in the liver tissue. Pbintoxication is also connected to the increased hepatic levels of lipid peroxidation and protein carbonylation and serum marker enzymes. But post treatment of animals with mangiferin after Pb exposure could change the alterations in the activities of the antioxidant enzymes and the level of GSH. It also modulated the levels of lipid peroxidation and protein carbonylation and serum marker enzymes. We studied Pb induced mode of cell death and its protection by mangiferin using DNA fragmentation and flowcytometric analyses. DNA fragmentation is one of the most often used techniques in the study of cell death. Internucleosomal DNA fragmentation can be visualized by gel electrophoresis as 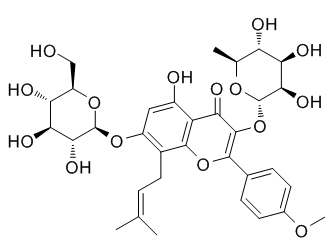 the characteristic DNA ladder formation and was considered as a biochemical hallmark of apoptosis. In our study, DNA gel electrophoresis showed that Pb exposure caused DNA fragmentation which appeared as a ladder in the agarose-ethidium bromide gel. The result of this study clearly suggests that Pb induced cell death occurred via apoptotic pathway. Mangiferin could, however, inhibit the Pb induced DNA fragmentation and apoptotic cell death. Flowcytometric analyses also demonstrated that Pb mostly damaged hepatocytes via apoptotic pathway. Simultaneous treatment with mangiferin, on the other hand, decreased the degree of Pbinduced apoptotic cell death. Multicellular organisms have three well-characterized subfamilies of mitogen activated protein kinases. The members of the family are basically serine/ threonine kinases, activated by dual phosphorylation on their threonine and tyrosine residues and are projected as critical redox signaling proteins.
the characteristic DNA ladder formation and was considered as a biochemical hallmark of apoptosis. In our study, DNA gel electrophoresis showed that Pb exposure caused DNA fragmentation which appeared as a ladder in the agarose-ethidium bromide gel. The result of this study clearly suggests that Pb induced cell death occurred via apoptotic pathway. Mangiferin could, however, inhibit the Pb induced DNA fragmentation and apoptotic cell death. Flowcytometric analyses also demonstrated that Pb mostly damaged hepatocytes via apoptotic pathway. Simultaneous treatment with mangiferin, on the other hand, decreased the degree of Pbinduced apoptotic cell death. Multicellular organisms have three well-characterized subfamilies of mitogen activated protein kinases. The members of the family are basically serine/ threonine kinases, activated by dual phosphorylation on their threonine and tyrosine residues and are projected as critical redox signaling proteins.
Limited developmental potential of Dnmt1 null ESCs and to impaired survival or proliferation of their differentiated progeny
These transcription factors establish a core network that, in cooperation with epigenetic modifiers, non coding RNAs and the c-Myc transcriptional network, orchestrates the pluripotency expression program and suppresses differentiation programs. Recent data suggest that the same core pluripotency factors also play crucial roles in initial cell fate choices. Differentiation signals directly modulate Oct4 and Sox2 protein levels, leading to changes in their genome wide binding profiles and thus initiating lineage selection without prior activation of lineage specification factors. By indexing chromatin states through DNA and histone modification, epigenetic factors ensure stable propagation of transcription programs and thus contribute to cell identity. At the same time, epigenetic marks are typically reversible and the enzymatic systems that set and erase them respond directly or indirectly to environmental signals, providing the necessary plasticity for the dynamic changes of transcription programs 3,4,5-Trimethoxyphenylacetic acid required for progressive differentiation. Although many epigenetic factors and chromatin remodelers have a role in stabilizing the pluripotent ESC state, most are actually not strictly required for its establishment and/or maintenance. This property and the demonstration that ESC self renewal is minimally dependent on extrinsic signaling have led to the concept that the pluripotent state of na? ��ve epiblast and ESCs represents a ground proliferative state relatively independent from epigenetic regulation. In contrast, most epigenetic regulators are required for proper execution of transcriptional programs driving lineage commitment and progression of differentiation. In mammals DNA methylation plays major roles in the control of gene expression 4-(Benzyloxy)phenol during development and differentiation. The importance of DNA methylation for proper development is underscored by the embryonic lethal phenotypes of mice lacking major DNA methyltransferases. These as well as many other studies have established that Dnmt3a and Dnmt3b, together with the catalytically inactive co-factor Dnmt3L, set DNA methylation patterns during embryogenesis and gametogenesis, while Dnmt1 is mainly responsible for maintaining these patterns through cell replication. However, further studies have shown that Dnmt3 enzymes are also required for long term maintenance of DNA methylation patterns and for their dynamic modulation in processes other than development. In embryos with homozygous inactivation of single Dnmt genes as well as in double Dnmt3a and 3b null embryos development arrests well after gastrulation, clearly showing that these enzymes are dispensable for the formation of na? ��ve epiblast. In addition, corresponding Dnmt null ESCs can be readily derived from blastocysts or by direct gene targeting even in the case of triple knockout of all catalytically active Dnmts. Dnmt1 null ESCs have about 20% residual genomic methylation, mostly located in repetitive sequences. TKO ESCs exhibit complete loss of genomic methylation, clearly showing that neither Dnmts nor DNA methylation are required for survival and self renewal of ESCs. Restoring Dnmt1 expression rescues the ability of Dnmt1 null ESCs to form teratomas, contribute to chimeras and complement tetraploid embryos.  Analogously, the ability to form teratomas is restored in Dnmt3 DKO ESCs upon expression of either Dnmt3 protein. Therefore, functional pluripotency of ESCs is not permanently compromised by the loss of either Dnmt1 or Dnmt3 proteins and consequent loss of DNA methylation.
Analogously, the ability to form teratomas is restored in Dnmt3 DKO ESCs upon expression of either Dnmt3 protein. Therefore, functional pluripotency of ESCs is not permanently compromised by the loss of either Dnmt1 or Dnmt3 proteins and consequent loss of DNA methylation.