The differences observed between our data and those in literature could suggest a different role of salicylic acid in TYLCSV infection but they could also be explained considering that. Autophagy is an evolutionarily conserved mechanism for intracellular recycling whereby large protein complexes and aggregates, organelles, and even invading pathogens are encapsulated in specific vesicles called autophagosomes, which are then engulfed and digested by vacuoles/lysosomes. This process is essential for tissue homeostasis and development, TAPI-0 and plays a critical role in the ability of plants to survive nutrient starvation as well as to exposure to abiotic and biotic stresses. The analysis of overrepresented functional categories highlighted a significant enrichment of autophagy-related transcripts among the genes up-regulated during the TYLCSV/tomato compatible interaction. All these transcripts were related to the formation of autophagosomes. In particular, several transcripts coding for isoforms of ATG8, key regulator and marker of the autophagic process, were induced during infection. ATG8 is an ubiquitin-like protein required for the formation of autophagosomal membranes. During the autophagic process, ATG8 anchors to the autophagosomes membranes by a carboxyl terminal phosphatidylethanolamine lipid. Interestingly, activation of ATG8 genes was also observed in Arabidopsis infected by CabLCV. Other TYLCSV-induced transcripts coded for ATG9, involved in the membrane delivery to autophagosomes, and ATG12, an ubiquitin-like protein involved in the formation of the ATG8-PE complex. Being intracellular parasites, viruses deal with the autophagic machinery during the infection process. In this respect, autophagy can play both anti-viral and pro-viral roles in viral life cycle and pathogenesis. On one hand, autophagy proteins can target viral components for lysosomal degradation and play a role in initiating immune response to infection. In this context, we can suppose that the observed up-regulation of autophagy genes is a host attempt to counteract viral invasion by elimination of exogenous viral particles. On the other hand, some animal viruses are able to hijack autophagy to foster their intracellular growth. In plants it is known that virus multiplication sequesters important cellular components, TAPI-2 causing cell starvation and activation of genes involved in programmed cell death. Cell death would be detrimental to viruses, in particular those like TYLCSV, which are restricted to a few cells in the phloem. Activation of autophagy by negatively regulating PCD, would prevent cells to die, thus maintaining a favourable environment for the virus. Clinical and experimental results have shown that the peripheral analgesic efficacy of opioids is enhanced in the presence of inflammation and tissue injury. The mechanisms responsible for this phenomenon involves the following events: increases in the opioid receptor mRNA level and opioid receptor expression level, an increase in the axonal transport and accumulation of these receptors in the peripheral sensory nerve endings, and an increase in the opioid agonist binding to their corresponding receptor. Moreover, inflammation induces the release of endogenous opioid peptides. Studies investigating the molecular mechanisms involved in the enhanced effects of opioid treatments in the periphery caused by inflammation mainly focus on the m-opioid receptor and the chronic inflammatory processes. Inflammation enhances the axonal transport of opioid receptors toward the periphery, which is preceded by an increase in opioid receptor mRNA transcription.
Author: targets inhibitor
Many novel proteins and transcripts with hitherto unknown links to FGF21 signal transduction pathways
In an independent in vivo validation study, immunoassays were performed using commercially available antibodies against 5 proteins and mouse blood taken at the 24 h and 48 h time points. Consistent with the gene expression data, Ccl11 Kinase Inhibitor Library abmole bioscience protein levels were increased. The lack of increase in circulating Ccl11 at 24 hours post FGF21 treatment may reflect the specific differences in the kinetics of protein synthesis and/or secretion. Il1rn protein levels were significantly decreased at the 24 h time point following the high dose of PEG30-FGF21 Q108. This is in agreement with the microarray analysis where the Il1rn transcript was down-regulated by the 2.5 mg/kg dose of PEG30-FGF21 Q108 at the 2 day time point. The increase in Il1rn at 48 h post-dose may reflect feedback regulation, although this requires further experimentation. Immunoassays for the three other proteins did not show dose-dependent changes in the plasma after FGF21 treatment. There was, however, a significant decrease in the plasma levels of all three proteins at the highest dose of FGF21 tested at the 24 h time point in agreement with their transcript regulation. Further studies are required to fully validate these findings. In the context of secreted proteins, the insulin-sensitizing adipokine adiponectin is of particular relevance to FGF21 signaling. Two recent studies have shown that adiponectin is a downstream effector of FGF21. Furthermore, treatment with FGF21 enhanced both the expression and secretion of adiponectin in mouse adipocytes, and also increased the serum levels of adiponectin in mice. In the present study we did not see significant increases in adiponectin mRNA in adipose tissues in vivo. In parallel experiments, we treated 3T3L1 adipocytes with WT FGF21 at 0.3 and 1 ��g/ml and could not observe an increase in adiponectin protein in the media, despite increased Erk phosporylation, a robust indicator of FGF21 receptor activation. Moreover, rosiglitazone treatment did trigger a 65% increase in secreted adiponectin in the same experiment. Similarly, in mice treated with WT FGF21 a 2-fold increase in Erk phosphorylation, indicative of target engagement, was not associated with increased levels of plasma adiponectin after either 1 or 6 hours of treatment. Rosiglitazone was not used in this in vivo study since more chronic dosing has been  reported to be required to observe an increase in circulating adiponectin levels. Further experiments are needed to determine the basis of these apparent inconsistencies. Changes in the phosphorylation of downstream signaling molecules following FGF21 treatment in adipose/adipocytes are expected to be the most proximal effects of receptor activation, and could potentially be used as TE biomarkers in adipose tissue biopsies. We used an unbiased phosphoproteomic SILAC approach in 3T3L1 mouse adipocytes treated with FGF21 to identify such changes. As a quality control, LC-MS profiling of forward and reverse labeled samples demonstrated that the heavy isotope incorporation as well as the equal Reversine purchase mixing of heavy and light samples were of good quality. We subsequently identified and quantified a total of 1186 phosphopeptides corresponding to 671 unique proteins from 3T3L1 adipocytes. Among them, 137 phosphopeptides showed a greater than 1.5 fold change between vehicle and FGF21 treatment. Of these phosphorylation events, 127 were up-regulated while 10 were down-regulated by FGF21 treatment compared to vehicle. Enriched canonical pathways from the Ingenuity Pathway Analysis tool included Insulin Receptor Signaling, IGF-1 Signaling, and Phospholipase C Signaling. The aim of the present study was to further interrogate the signaling events downstream of FGF21 receptor activation and, as an added bonus we identified a robust panel of target engagement biomarkers that may be useful for clinical development of FGF21-based therapeutics.
reported to be required to observe an increase in circulating adiponectin levels. Further experiments are needed to determine the basis of these apparent inconsistencies. Changes in the phosphorylation of downstream signaling molecules following FGF21 treatment in adipose/adipocytes are expected to be the most proximal effects of receptor activation, and could potentially be used as TE biomarkers in adipose tissue biopsies. We used an unbiased phosphoproteomic SILAC approach in 3T3L1 mouse adipocytes treated with FGF21 to identify such changes. As a quality control, LC-MS profiling of forward and reverse labeled samples demonstrated that the heavy isotope incorporation as well as the equal Reversine purchase mixing of heavy and light samples were of good quality. We subsequently identified and quantified a total of 1186 phosphopeptides corresponding to 671 unique proteins from 3T3L1 adipocytes. Among them, 137 phosphopeptides showed a greater than 1.5 fold change between vehicle and FGF21 treatment. Of these phosphorylation events, 127 were up-regulated while 10 were down-regulated by FGF21 treatment compared to vehicle. Enriched canonical pathways from the Ingenuity Pathway Analysis tool included Insulin Receptor Signaling, IGF-1 Signaling, and Phospholipase C Signaling. The aim of the present study was to further interrogate the signaling events downstream of FGF21 receptor activation and, as an added bonus we identified a robust panel of target engagement biomarkers that may be useful for clinical development of FGF21-based therapeutics.
The peptides with a positively charged harboring a hydrophobic directly in the plant system
Since many peptides of group IV have a high hemolytic activity, we selected peptides of group I and group III for further studies. The most promising candidates are derivatives of SP1 and SP10. These peptides were highly active against a broad spectrum of bacteria, but CP-690550 showed low hemolytic activity. First, we analysed the toxicity of these peptides for plant cells by incubation with plant protoplast. All three peptides showed a very low phytotoxicity to plant protoplasts and therefore seem to be well suited as a plant protecting agent. Hints to cell death are loss of spherical shape, chloroplast release and agglomeration of protoplasts. Bacteria, fungi and viruses can dramatically affect yield and quality of crop plants, which can have enormous economic consequences. But the damage is not only due to loss of money. Moreover, some of these phytopathogenic Niraparib PARP inhibitor microorganisms can produce toxins, which can affect health of the consumers. For instance aflatoxins, contaminants produced by the fungi Aspergillus flavus and Aspergillus parasiticus in a variety of food crops, are known to cause human liver cancer, to affect growth of human and animals and to be immunosuppressive. Therefore, the control of the plant pathogens is not only important from the economic point of view, but also very important for human and animal health. The control of the pathogenic microorganisms relies mainly on chemical pesticides. However, many countries have undertaken regulatory changes in pesticide registration requirements with the aim of retaining only compounds being 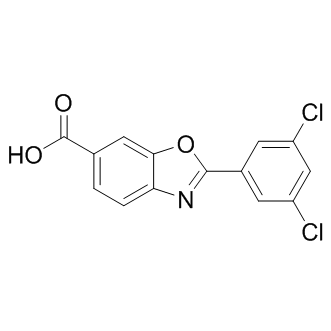 more selective, with lower toxicity and reduced negative environmental impact. The use of AMPs in agriculture was already proposed along with their discovery in the early 1980s. To date, nearly thousand natural antimicrobial peptides have been isolated from different organisms, such as mammals, insects, amphibians, and plants and can be found in AMP databases. The use of AMPs in animal or human medicine and plant protection may offer new possibilities to control microbial diseases that are still challenging to combat. However, most natural occurring AMPs exhibit a narrow activity spectrum, low activity against important pathogens or high toxicity against human and plant cells. To overcome these problems different types of antimicrobial peptides were designed. We developed helical antimicrobial peptides harbouring positively charged and hydrophobic clusters. In a first approach 16 different peptides with a length of 12 and 20 amino acids were developed. All designed peptides have a positive net charge of at least 3.76 at physiological pH, as it is the case for most of the natural occurring AMPs. SP1 and SP2 differ mainly in their N-terminal half and showed a broad antibacterial activity in a concentration range of 1�C2 mg/ml. Interestingly, SP3 and SP4 displayed a significant lower antibacterial activity, although the arrangement of hydrophobic and charged amino acid residues is the same as in SP1 and SP2. Notably, SP1 contains no alanine, whereas SP2, SP3 and SP4 contain two, three and four alanine residues, respectively. Wang et al. analyzed the amino acid composition of AMPs from different kingdoms of life listed in The Antimicrobial Peptide Database and determined frequently occurring residues. The most abundant residues in bacterial peptides are glycine and alanine. Possibly, with increasing alanine content the designed peptides reflect more analogy to bacterial peptides resulting in lower antimicrobial activity. Another difference among the peptides of group I is the slightly lower hydrophobicity of SP3 and SP4. This might affect the possibility of SP3 and SP4 to interact with bacterial membranes or to adopt membrane-induced amphipathicity at the expense of inhibiting bacterial growth.
more selective, with lower toxicity and reduced negative environmental impact. The use of AMPs in agriculture was already proposed along with their discovery in the early 1980s. To date, nearly thousand natural antimicrobial peptides have been isolated from different organisms, such as mammals, insects, amphibians, and plants and can be found in AMP databases. The use of AMPs in animal or human medicine and plant protection may offer new possibilities to control microbial diseases that are still challenging to combat. However, most natural occurring AMPs exhibit a narrow activity spectrum, low activity against important pathogens or high toxicity against human and plant cells. To overcome these problems different types of antimicrobial peptides were designed. We developed helical antimicrobial peptides harbouring positively charged and hydrophobic clusters. In a first approach 16 different peptides with a length of 12 and 20 amino acids were developed. All designed peptides have a positive net charge of at least 3.76 at physiological pH, as it is the case for most of the natural occurring AMPs. SP1 and SP2 differ mainly in their N-terminal half and showed a broad antibacterial activity in a concentration range of 1�C2 mg/ml. Interestingly, SP3 and SP4 displayed a significant lower antibacterial activity, although the arrangement of hydrophobic and charged amino acid residues is the same as in SP1 and SP2. Notably, SP1 contains no alanine, whereas SP2, SP3 and SP4 contain two, three and four alanine residues, respectively. Wang et al. analyzed the amino acid composition of AMPs from different kingdoms of life listed in The Antimicrobial Peptide Database and determined frequently occurring residues. The most abundant residues in bacterial peptides are glycine and alanine. Possibly, with increasing alanine content the designed peptides reflect more analogy to bacterial peptides resulting in lower antimicrobial activity. Another difference among the peptides of group I is the slightly lower hydrophobicity of SP3 and SP4. This might affect the possibility of SP3 and SP4 to interact with bacterial membranes or to adopt membrane-induced amphipathicity at the expense of inhibiting bacterial growth.
Cell system to analyze the consequences of in their structural organization and function
We therefore tested HspB1 and HspB5 level of phosphorylation in Neo, WT and R120G cells exposed or not to 60 mM menadione for 2 h, a duration that corresponded to the maximal level of phosphorylation induced by this drug. It is seen in Fig. 4A and 4Ba,b that, in non-treated cells, expression of wild type or mutant HspB5 did not significantly change the level of phosphorylation of HspB1 as demonstrated by calculating the R120G/WT ratio, which is representative of the modulation of the phosphorylation of a define serine site in response to the mutation. In contrast to HspB1, the phosphoserine sites of HspB5 were stimulated by the R120G mutation. In response to menadione treatment, the phosphorylation of HspB1 serine sites was increased by about 4-fold. In the case of HspB5, the effect was less intense and showed a decreased intensity depending on the N-terminal position of the serine sites. However, in oxidative conditions the R120G mutation still strongly enhanced HspB5 phosphorylation. A similar observation was made when cells were exposed to hydrogen peroxide treatment. Phosphorylation was Nutlin-3 further analyzed following cell fractionation in a 10,0006g supernatant and pellet. As expected, in non-treated Neo and WT cells, all the phosphorylated proteins were recovered in the soluble fraction. Analysis of 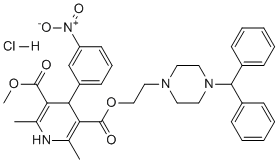 the fraction of HspB1 and HspB5 PLX-4720 Raf inhibitor present in the pellet fraction of R120G cells was performed by comparing, in the immunoblots, the ratios between the signals given by the percentage of the phosphorylated protein in the pellet to that of the percentage of the protein in that particular fraction. In non-treated R120G cells, the fraction of HspB1 in the pellet fraction had a phosphorylation index close to 1.0 while that of mutant HspB5 was in the range, depending on the serine site, of 0.2 to 0.5. This indicates that mutant HspB5 in the pellet fraction is less phosphorylated than its soluble counterpart and that HspB1 level of phosphorylation is less affected by the redistribution of this protein in the pellet fraction. Analysis of menadione treated Neo cells first showed that a large fraction of the total cellular content of HspB1 was recovered in the pellet fraction. The phenomenon was less intense in WT cells and more drastic in R120G cells. A similar observation was made for HspB5 in WT and R120G cells. Hence, the presence of these proteins in the pellet fraction correlated with the different sensitivity of these cells to oxidative stress. In response to menadione, HspB1 phosphorylation in Neo and WT cells was roughly proportional to the level of HspB1 in the fractions while it was slightly decreased in R120G cells. In contrast, HspB5 in WT cells showed a preferential phosphorylation in the pellet fraction. The phenomenon was not observed in R120G cells since the high level of menadione-induced phosphorylation was close to be proportional to the level of HspB5 present in the soluble and pellet fraction. To study the interaction between HspB1 and HspB5 in a defined cellular environment, HeLa cells were used since they constitutively express a high level of HspB1 but do not, or only weakly, express other interacting sHsps, particularly HspB5 and HspB6, which are known to form hetero-oligomeric complexes with HspB1. Genetically modified cells that stably express similar levels of either wild type or R120G mutated HspB5 were obtained. Of interest, HspB5 level of expression was close to that of endogenous HspB1. Another interesting characteristic was that more than 60% of HspB5 mutant expressed in these cells was recovered in a soluble form. This rather low level of aggregation suggests that the selected clones are probably adapted to the presence of the mutant protein. Indeed, R120G HspB5 polypeptide is well known for its drastic aggregation prone property; a phenomenon particularly intense in cells devoid of HspB1 expression.
the fraction of HspB1 and HspB5 PLX-4720 Raf inhibitor present in the pellet fraction of R120G cells was performed by comparing, in the immunoblots, the ratios between the signals given by the percentage of the phosphorylated protein in the pellet to that of the percentage of the protein in that particular fraction. In non-treated R120G cells, the fraction of HspB1 in the pellet fraction had a phosphorylation index close to 1.0 while that of mutant HspB5 was in the range, depending on the serine site, of 0.2 to 0.5. This indicates that mutant HspB5 in the pellet fraction is less phosphorylated than its soluble counterpart and that HspB1 level of phosphorylation is less affected by the redistribution of this protein in the pellet fraction. Analysis of menadione treated Neo cells first showed that a large fraction of the total cellular content of HspB1 was recovered in the pellet fraction. The phenomenon was less intense in WT cells and more drastic in R120G cells. A similar observation was made for HspB5 in WT and R120G cells. Hence, the presence of these proteins in the pellet fraction correlated with the different sensitivity of these cells to oxidative stress. In response to menadione, HspB1 phosphorylation in Neo and WT cells was roughly proportional to the level of HspB1 in the fractions while it was slightly decreased in R120G cells. In contrast, HspB5 in WT cells showed a preferential phosphorylation in the pellet fraction. The phenomenon was not observed in R120G cells since the high level of menadione-induced phosphorylation was close to be proportional to the level of HspB5 present in the soluble and pellet fraction. To study the interaction between HspB1 and HspB5 in a defined cellular environment, HeLa cells were used since they constitutively express a high level of HspB1 but do not, or only weakly, express other interacting sHsps, particularly HspB5 and HspB6, which are known to form hetero-oligomeric complexes with HspB1. Genetically modified cells that stably express similar levels of either wild type or R120G mutated HspB5 were obtained. Of interest, HspB5 level of expression was close to that of endogenous HspB1. Another interesting characteristic was that more than 60% of HspB5 mutant expressed in these cells was recovered in a soluble form. This rather low level of aggregation suggests that the selected clones are probably adapted to the presence of the mutant protein. Indeed, R120G HspB5 polypeptide is well known for its drastic aggregation prone property; a phenomenon particularly intense in cells devoid of HspB1 expression.
All HER family RTKs have been shown to be nuclear localized in its dimerization arm is innately positioned
This process leads to the activation of each receptors’ tyrosine kinase and the subsequent phosphorylation of tyrosine residues located on their C-terminal tails. Phosphorylated tyrosine residues recruit various intracellular adaptor and effector molecules that result in the propagation of growth promoting signal transduction cascades. Membrane-bound HER receptors WY 14643 activate numerous tumor promoting signaling cascades via this mechanism, including the PI3K/AKT, Ras/Raf/Mek/Erk, PLCc/PKC, and signal transducer and activator of transcription pathways. While the classical membrane-bound functions of HER family RTKs have been extensively studied, accumulating data suggest that these receptors can be found in the cell��s nucleus where they can function as co-transcriptional activators. To date, EGFR, HER2, and a nuclear variant of HER3 have been shown to function as co-transcriptional activators for cyclin D1. Clinically, nuclear EGFR has been correlated with poor overall survival in breast, ovarian, oropharyngeal, and gallbladder cancers. Nuclear EGFR has also been shown to play a role in resistance to numerous cancer therapies, including radiation, cisplatin, and the anti-EGFR therapies cetuximab and gefitinib. Collectively, these pivotal studies suggest that nuclear HER family receptors may enhance the tumorigenic phenotype of cancer cells, and therefore their nuclear roles must be further elucidated. The HER3 receptor has recently come to the forefront as playing a key role in HER family driven cancers. HER3 is a unique HER family member in that it has diminished tyrosine kinase activity due to the lack of specific amino acids within its kinase domain. However, HER3 plays a crucial role as an allosteric activator of other HER family members in addition to functioning as a signaling substrate through the direct recruitment and activation of PI3K. With the discovery of the various functions of nuclear EGFR and HER2, recent interest has prompted the investigation of nuclear HER3. HER3 was first identified to be nuclear localized in normal and malignant MG132 Proteasome inhibitor mammary epithelial cells in 2002. HER3 was also shown to be prominently nuclear localized in malignant prostate cancer tissues, where it was correlated with risk of disease progression. Most recently, two nuclear C-terminal splice variants of HER3 were identified and shown to function as co-transcriptional activators. Thus, the functions of nuclear HER3 are just beginning to unfold. In the current study we focused on identifying the amino acid regions on the C-terminal tail of HER3 that function as transactivation domains. First, numerous cancer cell lines were characterized for the expression of HER3, which was prominently nuclear localized in its full-length form. Next, various HER3 intracellular cytoplasmic domain regions fused to  the Gal4 DNA binding domain were analyzed, and the Cterminal domain of HER3 was shown to activate the Gal4 upstream activation sequence fused to luciferase. To identify specific C-terminal TADs, we created nine HER3-CTD truncation mutants and identified a bipartite region of 34 and 27 amino acids in length that contained the majority of HER3��s transactivation potential. To determine if the identified B1 and B2 TADs could augment nuclear HER3’s transcriptional function in cells, we first identified that full-length nuclear HER3 could associate and activate a 122 bp region of the cyclin D1 promoter when overexpressed. Importantly, the overexpression of HER3 lacking the B1 and B2 regions was severely hindered both in activating the 122 bp region of the cyclin D1 promoter and in enhancing transcription from the endogenous cyclin D1 gene. Collectively, these data suggest that this bipartite region of HER3 functions as a prominent TAD to mediate HER3’s nuclear functions.
the Gal4 DNA binding domain were analyzed, and the Cterminal domain of HER3 was shown to activate the Gal4 upstream activation sequence fused to luciferase. To identify specific C-terminal TADs, we created nine HER3-CTD truncation mutants and identified a bipartite region of 34 and 27 amino acids in length that contained the majority of HER3��s transactivation potential. To determine if the identified B1 and B2 TADs could augment nuclear HER3’s transcriptional function in cells, we first identified that full-length nuclear HER3 could associate and activate a 122 bp region of the cyclin D1 promoter when overexpressed. Importantly, the overexpression of HER3 lacking the B1 and B2 regions was severely hindered both in activating the 122 bp region of the cyclin D1 promoter and in enhancing transcription from the endogenous cyclin D1 gene. Collectively, these data suggest that this bipartite region of HER3 functions as a prominent TAD to mediate HER3’s nuclear functions.