Our proteomic analysis revealed that several enzymes of the citrate cycle and of oxidative phosphorylation were down-regulated in GC cells. These data show possible alterations in mitochondrion Cinoxacin function and a shift in energy production in the present GC cells, suggesting the Warburg effect. Proliferating tumor cells reprogram their metabolic pathways to generate energy and, thus, support the rapid cell division under stressful metabolic conditions that are characteristic of the abnormal tumor microenvironment. Even under normal oxygen concentrations, tumor cells shift from ATP generation through oxidative phosphorylation to ATP generation through glycolysis, converting most incoming glucose to lactate. It has been proposed that highly active glycolysis provides a biosynthetic advantage for tumor cells. Glycolysis provides enough metabolic intermediates by avoiding the oxidation of glucose, which is essential for the synthesis of macromolecules, such as lipids, proteins, and nucleic acids, during cell division. The lactate dehydrogenase and pyruvate dehydrogenase complexes control the metabolism of pyruvic acids, transforming to either lactic acids or acetyl-CoA 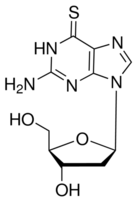 then entering the citrate cycle. The down-regulation of subunits of the LDH and PDH complexes suggests a reduction in pyruvate flux into the citrate cycle and a decrease in the rate of oxidative phosphorylation and oxygen consumption, reinforcing the glycolytic phenotype. Other down-regulated proteins, such as LIPF and GOT1, highlight the activation of other metabolic pathways with the impairment of the citrate cycle and oxidative phosphorylation. Several metabolic alterations that we observed in noncardia GC were also described by Cai et al. in a cardia GC proteomic study, suggesting that these metabolic alterations are not specific to a GC subtype based on tumor location. By the PANTHER system, the most significantly enriched pathway is the p53 pathway observed in the comparison between tumor and control samples. Additionally to its function in the DNA damage response and apoptosis, p53 is also a regulator of cell metabolism. p53 promotes oxidative phosphorylation and also inhibits the glycolytic pathway by up-regulating the expression of TP53-induced glycolysis and the apoptosis regulator. Therefore, the loss of p53 contributes to the acquisition of glycolytic phenotype. The loss of TP53 locus is a common finding in GC of individuals from Northern Brazil. Moreover, 20% of the GC analyzed by 2-DE presented p53 immunoreactivity. The p53 immunoreactivity usually depends on accumulation of mutated proteins in the cell, which leads to a longer half-life. The top networks of molecular interactions and functions were also identified using the IPA software. We showed several differently regulated proteins involved in cellular assembly and organization, and in inflammatory processes. The cellular assembly and organization was the principally enriched network observed in the comparison between controls and tumors with lymph node metastasis. Therefore, our data reveal that the molecules of the described subnetwork are important to the process of metastasis in noncardia Lomitapide Mesylate gastric carcinogenesis. Previous studies have demonstrated that GC is strongly linked to chronic inflammation, and that infection with H. pylori may trigger the chronic inflammation that can lead to malignancy. However, the exact mechanism of this process is still not known. The identified proteins add new pieces to this process in gastric carcinogenesis. The unsupervised hierarchical clustering of the differentially expressed proteins revealed that the tumors and control samples do not form two distinct separate clusters. Although, hierarchical clustering revealed one group composed by only controls, the other group presented all tumor samples and two misclassified control samples.
then entering the citrate cycle. The down-regulation of subunits of the LDH and PDH complexes suggests a reduction in pyruvate flux into the citrate cycle and a decrease in the rate of oxidative phosphorylation and oxygen consumption, reinforcing the glycolytic phenotype. Other down-regulated proteins, such as LIPF and GOT1, highlight the activation of other metabolic pathways with the impairment of the citrate cycle and oxidative phosphorylation. Several metabolic alterations that we observed in noncardia GC were also described by Cai et al. in a cardia GC proteomic study, suggesting that these metabolic alterations are not specific to a GC subtype based on tumor location. By the PANTHER system, the most significantly enriched pathway is the p53 pathway observed in the comparison between tumor and control samples. Additionally to its function in the DNA damage response and apoptosis, p53 is also a regulator of cell metabolism. p53 promotes oxidative phosphorylation and also inhibits the glycolytic pathway by up-regulating the expression of TP53-induced glycolysis and the apoptosis regulator. Therefore, the loss of p53 contributes to the acquisition of glycolytic phenotype. The loss of TP53 locus is a common finding in GC of individuals from Northern Brazil. Moreover, 20% of the GC analyzed by 2-DE presented p53 immunoreactivity. The p53 immunoreactivity usually depends on accumulation of mutated proteins in the cell, which leads to a longer half-life. The top networks of molecular interactions and functions were also identified using the IPA software. We showed several differently regulated proteins involved in cellular assembly and organization, and in inflammatory processes. The cellular assembly and organization was the principally enriched network observed in the comparison between controls and tumors with lymph node metastasis. Therefore, our data reveal that the molecules of the described subnetwork are important to the process of metastasis in noncardia Lomitapide Mesylate gastric carcinogenesis. Previous studies have demonstrated that GC is strongly linked to chronic inflammation, and that infection with H. pylori may trigger the chronic inflammation that can lead to malignancy. However, the exact mechanism of this process is still not known. The identified proteins add new pieces to this process in gastric carcinogenesis. The unsupervised hierarchical clustering of the differentially expressed proteins revealed that the tumors and control samples do not form two distinct separate clusters. Although, hierarchical clustering revealed one group composed by only controls, the other group presented all tumor samples and two misclassified control samples.
Author: targets inhibitor
Both GATA2 and AP1 binding sites are necessary for epithelial induction of ET-1 under hypoxia
By the evolutionarily conserved Hypoxia Inducible Factor family of basic helix-loop-helix transcription factors. HIFs are heterodimers of a beta subunit, and an alpha subunit. While ARNT levels are not sensitive to oxygen, both HIFa stability and its transcriptional activity are regulated by oxygen-dependent hydroxylation. Under oxygen restriction, HIFa subunits escape  proteasomal degradation, heterodimerize with HIFb subunits and translocate to the cell nucleus, where they bind the RCGTG consensus sequence within regulatory regions of target genes, leading to their transcriptional activation in hypoxia. Mammals present three isoforms of HIFa that differ in their tissue distribution, HIF1a being the more ubiquitous and best characterized. A large number of studies focusing on single genes have identified individual HIF targets that, collectively, account for the functional responses to hypoxia, mainly metabolic adaptation and induction of angiogenesis. More recently, works employing HIF1a and HIF2a chromatin immunoprecipitation coupled to genomic microarrays or high-throughput sequencing have addressed the genome-wide identification of HIF binding locations, thereby improving the existing knowledge on the HIF-modulated transcriptome and largely confirming the RCGTG HIF binding consensus. Additionally, these studies have provided important insights into the global properties of HIF1 binding and transactivation. First, these works reported a significant association between the presence of a HIF binding site and hypoxic induction of the neighboring genes. The same trend was not found for genes repressed by hypoxia, Folinic acid calcium salt pentahydrate suggesting that hypoxia-mediated repression is largely indirect or HIF independent. Furthermore, they have clearly shown that only a small subset of about a hundred of all RCGTG-containing genes is robustly regulated by hypoxia. Hence, and in agreement with work on other transcription factors, HIFs bind a small proportion of potential binding sites, albeit the basis of their binding and target selectivity are incompletely understood. Understanding the mechanisms that explain HIFs transactivation selectivity is of paramount importance to expand our knowledge on transcriptional regulation and to improve the sensitivity and specificity of genome-wide efforts to characterize the HIF transcriptional response. DNA accessibility of transcription factor binding sites can clearly contribute to binding selectivity. For HIFs, recent evidence includes enhanced HIF1 and HIF2 binding to normoxic DNAse hypersensitivity sites and enrichment of HIF1 binding in the proximity of genes with a “permissive” transcriptional state in normoxia, as evidenced by significant basal expression. Additionally, DNA methylation has been also shown to modulate HIF1 binding, as originally demonstrated for the 39 enhancer of the Lomitapide Mesylate erythropoietin gene. A further mechanism that can impact target selectivity is direct or indirect cooperativity between transcription factors. Models of direct cooperativity have been mainly derived from developmental enhancers, and include the strict enhanceosome model, where cooperative occupancy occurs through extensive protein-protein interactions between TFs or common cofactors, and the more flexible billboard model, which suggests that enhancers contain submodules that interact independently or redundantly with promoters. Conversely, indirect cooperativity is based on the equilibrium competition between nucleosomes and DNA-binding proteins, thereby not requiring protein-protein interactions. In the case of HIFmediated transcription, the binding of cooperating transcription factors has been demonstrated for several target genes. In particular, HIF-mediated expression of the erythropoietin gene requires an adjacent HNF4 binding site, and PAI-1 induction by hypoxia has been linked to cooperative promoter activation by CEBPa.
proteasomal degradation, heterodimerize with HIFb subunits and translocate to the cell nucleus, where they bind the RCGTG consensus sequence within regulatory regions of target genes, leading to their transcriptional activation in hypoxia. Mammals present three isoforms of HIFa that differ in their tissue distribution, HIF1a being the more ubiquitous and best characterized. A large number of studies focusing on single genes have identified individual HIF targets that, collectively, account for the functional responses to hypoxia, mainly metabolic adaptation and induction of angiogenesis. More recently, works employing HIF1a and HIF2a chromatin immunoprecipitation coupled to genomic microarrays or high-throughput sequencing have addressed the genome-wide identification of HIF binding locations, thereby improving the existing knowledge on the HIF-modulated transcriptome and largely confirming the RCGTG HIF binding consensus. Additionally, these studies have provided important insights into the global properties of HIF1 binding and transactivation. First, these works reported a significant association between the presence of a HIF binding site and hypoxic induction of the neighboring genes. The same trend was not found for genes repressed by hypoxia, Folinic acid calcium salt pentahydrate suggesting that hypoxia-mediated repression is largely indirect or HIF independent. Furthermore, they have clearly shown that only a small subset of about a hundred of all RCGTG-containing genes is robustly regulated by hypoxia. Hence, and in agreement with work on other transcription factors, HIFs bind a small proportion of potential binding sites, albeit the basis of their binding and target selectivity are incompletely understood. Understanding the mechanisms that explain HIFs transactivation selectivity is of paramount importance to expand our knowledge on transcriptional regulation and to improve the sensitivity and specificity of genome-wide efforts to characterize the HIF transcriptional response. DNA accessibility of transcription factor binding sites can clearly contribute to binding selectivity. For HIFs, recent evidence includes enhanced HIF1 and HIF2 binding to normoxic DNAse hypersensitivity sites and enrichment of HIF1 binding in the proximity of genes with a “permissive” transcriptional state in normoxia, as evidenced by significant basal expression. Additionally, DNA methylation has been also shown to modulate HIF1 binding, as originally demonstrated for the 39 enhancer of the Lomitapide Mesylate erythropoietin gene. A further mechanism that can impact target selectivity is direct or indirect cooperativity between transcription factors. Models of direct cooperativity have been mainly derived from developmental enhancers, and include the strict enhanceosome model, where cooperative occupancy occurs through extensive protein-protein interactions between TFs or common cofactors, and the more flexible billboard model, which suggests that enhancers contain submodules that interact independently or redundantly with promoters. Conversely, indirect cooperativity is based on the equilibrium competition between nucleosomes and DNA-binding proteins, thereby not requiring protein-protein interactions. In the case of HIFmediated transcription, the binding of cooperating transcription factors has been demonstrated for several target genes. In particular, HIF-mediated expression of the erythropoietin gene requires an adjacent HNF4 binding site, and PAI-1 induction by hypoxia has been linked to cooperative promoter activation by CEBPa.
Those in the set of core HIF binding regions by screening for evolutionarily conserved HIF binding consensus sequences
Other examples include cooperation with Smads, Sp1 or CREB. Additionally, USFs have been shown to complement HIF binding either at neighbouring or identical sites, while collaboration with ETS transcription factors has been proposed to play a role in HIF2a target selectivity. Recent genome-wide approaches relying on experimental and computational identification of HIF binding sites have reported overrepresented transcription factor binding sites in the flanking sequences that might be indicative of transcriptional cooperativity. However, significant differences exist in the overrepresented TFBSs predicted in each study, and the functional significance of these enriched motifs remains unclear. Gene expression profiling indicates that the expression of thousands of genes changes with hypoxia, with vast cell-type differences in the specific genes being regulated. HIF1a ChIP-chip binding Mechlorethamine hydrochloride locations have been reported in cell lines of diverse tissue origin, namely HepG2 hepatocarcinoma cells, MCF-7 breast cancer cells and U87 glioma cells, showing differences in the binding sites identified in each experiment. In previous studies we integrated microarray expression profiling experiments and HIF binding site predictions in a core set of tissue-independent HIF target genes. To further investigate the selectivity of HIF1 binding, in this work we conducted HIF1a ChIP-chip in cervical carcinoma HeLa cells and observed largely non-overlapping binding locations with previous studies. To explore the role of cooperativity in HIF target selection, we integrated HIF1 alpha ChIP-chip binding locations across cell-types with a meta-analysis of gene expression profiles of cells exposed to hypoxia. Computational prediction of enriched transcription factor binding sites in this integrated set suggested several stress-responsive transcription factors as potential HIF1 collaborators. Experimental validation of these predictions in cell-based reporter assays indicates that binding sites for stressresponsive transcription factors other than HIFs, such as CEBPs, contribute to cooperative hypoxic activation of individual targets. To obtain a set of background genomic regions, custom perl scripts were used to exploit the microarray metaanalysis results for the identification of genes harbouring conserved RCGTG motifs but that are unlikely to be modulated by hypoxia. To this end, gene loci that contained conserved RCGTG motifs in their non-coding sequences were first selected. For these genes, each of their probes was examined, and only genes for which all of their associated probes exhibited a mean fold value within 0.25 standard deviations of the global mean in 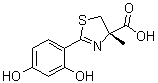 each of the 19 datasets employed in the meta-analysis were considered as not induced by hypoxia. The selected coordinates of conserved RCGTG motifs mapping to these loci were extended as previously described for the set of core HIF binding sites. Genomic regions from this collection were further selected to match the frequency of genomic locations found in the core HBR set. Briefly, Perl scripts were used to annotate core HBRs as promoter, 59UTR, intronic or 39UTR genomic locations and to choose, from the whole collection, a random sample according to the proportions of genomic locations found in the core HBR set. Similarly as with the set of core HIF binding regions, multiple sequence alignments corresponding to the selected control regions were retrieved. The length of flanking non-coding sequences was based on evolutionary conservation, as Chlorhexidine hydrochloride indicated by genomic annotation of PhastCons elements. Statistical assessment of sequence motif enrichment in this set of sequences requires comparison with a background set, the election of this set greatly influencing the results of the analysis.
each of the 19 datasets employed in the meta-analysis were considered as not induced by hypoxia. The selected coordinates of conserved RCGTG motifs mapping to these loci were extended as previously described for the set of core HIF binding sites. Genomic regions from this collection were further selected to match the frequency of genomic locations found in the core HBR set. Briefly, Perl scripts were used to annotate core HBRs as promoter, 59UTR, intronic or 39UTR genomic locations and to choose, from the whole collection, a random sample according to the proportions of genomic locations found in the core HBR set. Similarly as with the set of core HIF binding regions, multiple sequence alignments corresponding to the selected control regions were retrieved. The length of flanking non-coding sequences was based on evolutionary conservation, as Chlorhexidine hydrochloride indicated by genomic annotation of PhastCons elements. Statistical assessment of sequence motif enrichment in this set of sequences requires comparison with a background set, the election of this set greatly influencing the results of the analysis.
Both intrauterine and postnatal malnutrition promote the same patterns of Ang II receptor expression
As discussed above, in a similar model of maternal undernutrition, plasma levels of the anorexigenic leptin were increased and levels of the orexigenic prolactin were decreased. In addition to decreasing maternal food intake, this hormone imbalance could have imprinted an impaired dietary behavior on the progeny. Evidence for this concerns elevated plasma levels of leptin in  pups from mothers that had been undernourished during lactation, as a result of direct transmission via milk. Acid maceration is not the gold-standard method for estimating the Pimozide number of nephrons. However, reproducibility of counting using Lomitapide Mesylate different kidneys and more than one independent observer, together with comparable control values with data from other laboratories, demonstrated a profound influence of programming in terms of the number of nephrons in adult kidneys. The reduced number of nephrons and the structural glomerular alterations demonstrate that the decreased kidney mass of programmed animals was not a simple relationship to lower body mass. The decreased number of nephrons and decreased capillary area with consequent hypofiltration could be associated with hyperfiltration in remnant healthy nephrons, where the increased intracapillary pressure would contribute to their late and progressive self-destruction with late onset hypertension. The onset of hypertension in older rats supports this view. An increase in the glomerular area would be expected when the number of nephrons is decreased, and compensatory hyperfiltration occurs in some remnant glomeruli. It could be that intense collagen deposition in the cortex of programmed animals affects many glomerular structures, causing a global reduction in the size of the Bowman’s capsule and glomerular capillary tufts, with preservation or an increase in the area of others. It is unexpected that GFR increases by 70% in 60 day-old programmed rats, despite reduced glomerular areas as presented in Fig. 4. However, this could be explained by exacerbated hyperfiltration in the preserved nephrons. The intense proteinuria that accompanies the increase in GRF and UNa reveals important and early damage in the filtration barrier that could evolve into global impairment of renal function in programmed rats. Therefore, kidneys from progeny that were programmed during lactation suffer from early severe morphological, and consequently functional, alterations in glomerular components, adding to the molecular alterations in proximal tubules, as discussed below. The results depicted in Figs. 6, 7, 8, 9 demonstrate that programming during lactation strongly imprints the molecular machinery responsible for the majority of Na + absorption and the fine tuning of this process, leading to augmented recovery of the filtered fluid, which can subsequently contribute to the onset of hypertension. It is interesting that placental undernutrition promotes different modifications from those observed in the present study of young adult rats. This supports the view that depending on the window of development, undernutrition may evoke different signals that affect the same organs in different ways. Up-regulation of AT1 receptors and down-regulation of AT2 receptors could represent the persistence into adulthood of an adaptative response towards impaired RAS functioning in early life. Lower local Ang II production in early life could be compensated by the programming-induced reciprocal alterations in the populations of AT1 and AT2 receptors as demonstrated in Fig. 10, which persisted into adult life. In contrast to the results concerning active Na + transporters.
pups from mothers that had been undernourished during lactation, as a result of direct transmission via milk. Acid maceration is not the gold-standard method for estimating the Pimozide number of nephrons. However, reproducibility of counting using Lomitapide Mesylate different kidneys and more than one independent observer, together with comparable control values with data from other laboratories, demonstrated a profound influence of programming in terms of the number of nephrons in adult kidneys. The reduced number of nephrons and the structural glomerular alterations demonstrate that the decreased kidney mass of programmed animals was not a simple relationship to lower body mass. The decreased number of nephrons and decreased capillary area with consequent hypofiltration could be associated with hyperfiltration in remnant healthy nephrons, where the increased intracapillary pressure would contribute to their late and progressive self-destruction with late onset hypertension. The onset of hypertension in older rats supports this view. An increase in the glomerular area would be expected when the number of nephrons is decreased, and compensatory hyperfiltration occurs in some remnant glomeruli. It could be that intense collagen deposition in the cortex of programmed animals affects many glomerular structures, causing a global reduction in the size of the Bowman’s capsule and glomerular capillary tufts, with preservation or an increase in the area of others. It is unexpected that GFR increases by 70% in 60 day-old programmed rats, despite reduced glomerular areas as presented in Fig. 4. However, this could be explained by exacerbated hyperfiltration in the preserved nephrons. The intense proteinuria that accompanies the increase in GRF and UNa reveals important and early damage in the filtration barrier that could evolve into global impairment of renal function in programmed rats. Therefore, kidneys from progeny that were programmed during lactation suffer from early severe morphological, and consequently functional, alterations in glomerular components, adding to the molecular alterations in proximal tubules, as discussed below. The results depicted in Figs. 6, 7, 8, 9 demonstrate that programming during lactation strongly imprints the molecular machinery responsible for the majority of Na + absorption and the fine tuning of this process, leading to augmented recovery of the filtered fluid, which can subsequently contribute to the onset of hypertension. It is interesting that placental undernutrition promotes different modifications from those observed in the present study of young adult rats. This supports the view that depending on the window of development, undernutrition may evoke different signals that affect the same organs in different ways. Up-regulation of AT1 receptors and down-regulation of AT2 receptors could represent the persistence into adulthood of an adaptative response towards impaired RAS functioning in early life. Lower local Ang II production in early life could be compensated by the programming-induced reciprocal alterations in the populations of AT1 and AT2 receptors as demonstrated in Fig. 10, which persisted into adult life. In contrast to the results concerning active Na + transporters.
Parameters and metrics for working specifically with EST transcriptomic sequences produced by 454 sequencers
It was our last choice of the tested assemblers, according to evaluation metrics used. We also verified that some software works better with different clustering measures, such as the Celera assembler, which more effectively maps all the assembled reads into clusters. Most researchers would agree that the problem of sequence assembly and consensus production is highly complex and has not yet been solved, although most papers in the field simply choose a software program and proceed with assembly without further evaluations. Because sequence clustering is dependent on the informational content of the original reads, we recommend that authors test the appropriateness of the software for their own data before commencing the Lomitapide Mesylate annotation and analysis of unigenes. Based on the number of unigenes found here, we suspect that we have sequenced most of the coding sequences expressed by Actinopus spp. and G. cancriformis in their spinning glands. The number of unigenes in the Unigene database for the tick I. scapularis suggests that a number of unigenes in excess of the 25,663 found for G. cancriformis may be the rule for the Chelicerata clade. This excess of unigenes cannot be explained simply by the small size of the NGS-generated unigenes, although many unigenes probably represent either 39 or the 59 portions of genes. Our inability to identify most of the G. cancriformis unigenes by BLAST analysis indicates a lack of interest in Chelicerata clade genomics. We tried to fix this knowledge gap in the present publication by identifying more than 30,000 new genes for this clade and making them available to the research community in the SRA database. The amazing genetic Butenafine hydrochloride repertoire of spiders with their highly specialized tissues, such as the spinning and venom glands, will help researchers make biotechnological breakthroughs in the coming years, as has been recently discussed in the specialized literature. Remarkably, we found the metalloproteases from the astacin family to be extremely significant in the G. cancriformis spinning gland transcriptome. Genes from this family, which still have unknown functions, corresponded to 1.8% of all transcripts sequenced in the spinning glands, a number approximately 3.6 times larger than the number of expressed spidroin genes. Moreover, among the gene families that have undergone major amplifications since the ancestral split between the Mygalomorphae and Araneomorphae clades, we found that this family of metal-dependent proteinases has been amplified 10 times in the genome of the Araneomorphae/Orbiculariae  clade. The observed proteolytic effects of metalloproteinases in the venom of some Araneomorphae spiders are probably unrelated to their high expression in the spinning glands. Moreover, until recently, this subfamily of the meprin metalloproteases was believed to exist only in vertebrates, although even more recently, it was reported in the horseshoe crab Limulus polyphemus, an organism from the subphylum Chelicerata. Our work, therefore, corroborates the evidence that this gene family is present in the Arthropoda subphylum studied here. Moreover, KOG data from 2003 have classified a number of these proteins in the genomes of invertebrates such as D. melanogaster and Caenorhabditis elegans. Meprins consist of a single, membrane-anchored member of the astacin family and have been shown to be implicated in a number of complex cellular processes in higher eukaryotes, such as cell migration, immune reactions, and tissue differentiation. A clue to their function may come from some works reporting their relationship to human collagen fibers.
clade. The observed proteolytic effects of metalloproteinases in the venom of some Araneomorphae spiders are probably unrelated to their high expression in the spinning glands. Moreover, until recently, this subfamily of the meprin metalloproteases was believed to exist only in vertebrates, although even more recently, it was reported in the horseshoe crab Limulus polyphemus, an organism from the subphylum Chelicerata. Our work, therefore, corroborates the evidence that this gene family is present in the Arthropoda subphylum studied here. Moreover, KOG data from 2003 have classified a number of these proteins in the genomes of invertebrates such as D. melanogaster and Caenorhabditis elegans. Meprins consist of a single, membrane-anchored member of the astacin family and have been shown to be implicated in a number of complex cellular processes in higher eukaryotes, such as cell migration, immune reactions, and tissue differentiation. A clue to their function may come from some works reporting their relationship to human collagen fibers.