We found that ZFNs undergo proteasomal degradation and that MG132 increases ZFN levels, leading to enhanced genetic modifications by the ZFNs. Our protein stability study should lay the foundation for the advancement of ZFN technology; furthermore, the identification of MG132 as a small molecule that increases ZFN function is expected to facilitate the use of ZFNs. Targeted genetic modification using ZFNs can enable targeted gene insertion, correction, disruption, chromosomal rearrangement, 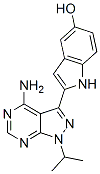 and regulatory region alteration. Gene editing using ZFNs is a promising technology as a powerful tool for studying biological processes and for the development of R428 advanced gene therapy to correct pathogenic genes. Here for the first time we investigated ZFN stability. Given that high levels of ZFN protein are associated with enhanced ZFN activity, our protein stability study should lay the foundation for further development of ZFN technology. Furthermore, ZFNs can be now delivered directly as protein and several doses of ZFN protein treatment are required to obtain sufficient ZFN activity, because the ZFNs are degraded within a few hours after treatment. Thus, the development of methods to maintain sufficient ZFN concentrations is important; our protein stability study should serve as a basis for this research. Even when a ZFN protein is continuously expressed by a DNA vector transfected into the target cells, inhibition of ZFN degradation increased the ZFN protein levels, leading to enhanced genetic modification. Porteus�� group has PB 203580 supply previously reported that short-term exposure to MG132 does not significantly increase the protein levels and activity of ZFNs that contain wild-type FokI nucleases. In contrast, we observed that MG132 treatment increased ZFN activity 2.4 or 2-fold. One possible reason for this discrepancy could be the different FokI nucleases employed in the two experiments: the ZFN used by Porteus�� group contained the wildtype FokI nuclease, whereas we used ZFNs with a modified FokI nuclease, which is improved from the wild type and is now predominantly used. This difference in the ZFN amino acid sequence might affect the rate of ZFN proteolysis. Another reason could be the difference in the MG132 concentration and duration of exposure: we treated cells with MG132 for 60 hours at 1, 2, and 5 mM, whereas Porteus�� group used 10 mM of MG132 for only 4 hours. We observed significantly decreased cell viability at 10 mM of MG132 when cells were treated for 60 hours. In addition, the application of MG132 to human embryonic stem cells caused cytotoxic effects even at very low dosage, which is compatible with the previous reports that showed similar cytotoxic effects of MG132 on hESCs. Here we showed that ZFN activity can be enhanced using a small molecule, MG132. To our knowledge, this is the first study reporting that a small molecule can regulate ZFN function. Identifying small molecules with this property is important given that ZFN technology is actively being studied as a tool for gene therapy and to analyze biological processes. Although MG132 is not a FDA-approved drug, other FDA-approved proteasomal inhibitors such as bortezomib might be used together with ZFNs to enhance the effect of gene therapy. Indeed, it has been recently reported that bortezomib can increase the effect of a ZFN-expressing adeno-associated virus.
and regulatory region alteration. Gene editing using ZFNs is a promising technology as a powerful tool for studying biological processes and for the development of R428 advanced gene therapy to correct pathogenic genes. Here for the first time we investigated ZFN stability. Given that high levels of ZFN protein are associated with enhanced ZFN activity, our protein stability study should lay the foundation for further development of ZFN technology. Furthermore, ZFNs can be now delivered directly as protein and several doses of ZFN protein treatment are required to obtain sufficient ZFN activity, because the ZFNs are degraded within a few hours after treatment. Thus, the development of methods to maintain sufficient ZFN concentrations is important; our protein stability study should serve as a basis for this research. Even when a ZFN protein is continuously expressed by a DNA vector transfected into the target cells, inhibition of ZFN degradation increased the ZFN protein levels, leading to enhanced genetic modification. Porteus�� group has PB 203580 supply previously reported that short-term exposure to MG132 does not significantly increase the protein levels and activity of ZFNs that contain wild-type FokI nucleases. In contrast, we observed that MG132 treatment increased ZFN activity 2.4 or 2-fold. One possible reason for this discrepancy could be the different FokI nucleases employed in the two experiments: the ZFN used by Porteus�� group contained the wildtype FokI nuclease, whereas we used ZFNs with a modified FokI nuclease, which is improved from the wild type and is now predominantly used. This difference in the ZFN amino acid sequence might affect the rate of ZFN proteolysis. Another reason could be the difference in the MG132 concentration and duration of exposure: we treated cells with MG132 for 60 hours at 1, 2, and 5 mM, whereas Porteus�� group used 10 mM of MG132 for only 4 hours. We observed significantly decreased cell viability at 10 mM of MG132 when cells were treated for 60 hours. In addition, the application of MG132 to human embryonic stem cells caused cytotoxic effects even at very low dosage, which is compatible with the previous reports that showed similar cytotoxic effects of MG132 on hESCs. Here we showed that ZFN activity can be enhanced using a small molecule, MG132. To our knowledge, this is the first study reporting that a small molecule can regulate ZFN function. Identifying small molecules with this property is important given that ZFN technology is actively being studied as a tool for gene therapy and to analyze biological processes. Although MG132 is not a FDA-approved drug, other FDA-approved proteasomal inhibitors such as bortezomib might be used together with ZFNs to enhance the effect of gene therapy. Indeed, it has been recently reported that bortezomib can increase the effect of a ZFN-expressing adeno-associated virus.
Month: August 2019
Comparing the respons fraction of ZFN-treated cells small molecules can be used in vitro to facilitate gene editing
In conclusion, we show that ZFN proteins have a relatively short half-life and that their turn-over is regulated by the UPP. Furthermore, treatment with the proteasome inhibitor MG132 blocked ZFN protein PF-4217903 cost degradation and extended its half-life, resulting in increased ZFN protein levels and enhanced genetic modification. Our protein stability study should lay the foundation for further development of ZFN technology. The identification of small molecules that increase ZFN protein levels will facilitate the application of ZFNs. To measure the level of functional miRNA in a manner that avoids detecting miRNA mimic trapped in non-functional locations, we immunoprecipitated UV cross-linked RISC from control and transfected cells and measured the amount of RISCassociated miR-200a by deep sequencing of the miRNA-sized RNA fraction in the immunoprecipitate. This revealed that the amount of RISC-associated miR-200a in the transfected cells was approximately equal to the level of other abundant miRNAs. This is proportionally much less than the level of miR200a measured by qPCR, indicating most of the transfected miRNA mimic is not bound to Argonaute and consequently is not functional. Similar results were obtained following transfection of a different miRNA, miR-200b. Thus, although qPCR is a valid technique to measure total miRNA amount, this can be very different from the amount of functional miRNA. Given the majority of miRNA mimic detected by qPCR did not represent the active Argonaute-bound population, we determined its sub-cellular localisation by transfecting a fluorescent siRNA and examining the transfected cells by fluorescence microscopy. The majority of the siRNA did not co-localise with Argonaute, which is consistent with earlier reports of transfected siRNA localising in large cytoplasmic aggregates that are distinct from the GW bodies that are known for their role in RNA silencing. Instead the vast majority of miRNA transfected with either HiPerfect,, RNAi-Max or Lipofectamine 2000 localised with or adjacent to lysosomes, matching earlier reports of lipid-based siRNA transfection. Therefore, the high level of transfected miRNA detected by qPCR is largely attributable to their retention within vesicles and subsequent amplification by qPCR following lysis. Hence, the use of qPCR to measure a miRNA after transient transfection can give the false impression that the miRNA is at massively nonphysiological level, whereas the amount of miRNA bound to Argonaute may indeed be appropriately physiological. On the other hand, it is conceivable that an inefficient  transfection that results in just a small proportion of cells being transfected could appear to produce an adequate level of miRNA, if measured by qPCR. It is more appropriate to use an assay of miRNA function to verify the effectiveness of the transfection. Of additional interest to users of miRNA mimics for transient transfection, we were able to confirm from our sequencing of the Argonaute-bound pool of small RNAs, that while a miRNA mimic with unmodified passenger strand results in abundant incorporation of the passenger strand into RISC, raising the potential for extensive off-target effects, a mimic that is modified to limit the incorporation of the passenger strand into RISC does indeed achieve this. Although the merits of modified mimics have been previously recognised, published evidence for this is NVP-BKM120 limited to date and has been based largely on reporter assays.
transfection that results in just a small proportion of cells being transfected could appear to produce an adequate level of miRNA, if measured by qPCR. It is more appropriate to use an assay of miRNA function to verify the effectiveness of the transfection. Of additional interest to users of miRNA mimics for transient transfection, we were able to confirm from our sequencing of the Argonaute-bound pool of small RNAs, that while a miRNA mimic with unmodified passenger strand results in abundant incorporation of the passenger strand into RISC, raising the potential for extensive off-target effects, a mimic that is modified to limit the incorporation of the passenger strand into RISC does indeed achieve this. Although the merits of modified mimics have been previously recognised, published evidence for this is NVP-BKM120 limited to date and has been based largely on reporter assays.
We wished to check ZFN proteolysis with MG132 and determine the effects on mediated gene disruption
For example, the ZFN nuclease domain has been modified to improve ZFN activity and Dinaciclib CDK inhibitor specificity. Additionally, modifying the culture temperature caused a significant increase in ZFN activity. Furthermore, our group recently reported a simple method to enrich cells that contain ZFN-induced gene disruptions. Given that these simple methods to improve the ZFN function have facilitated the use of ZFNs, the identification of small molecules that increase ZFN function should likewise efficiently facilitate the application of ZFNs. However, such small molecules have yet to be identified. It has been observed that ZFN protein levels are directly correlated with ZFN function. Culturing the cells at low temperature increases ZFN function at least in part because ZFN protein levels increase. We also observed that cell populations that are enriched with gene-disrupted cells have high ZFN levels as compared to control cells. Recently, direct delivery of ZFN proteins has been shown to be safer associated with negligible offtarget effects. These ZFN proteins could penetrate the cells without any additional cell-penetrating peptide sequences and were able to transduce into several cell types including those that are hard to transfect. However, due to degradation of the delivered protein, it was necessary to treat the cells several times with the ZFN protein to obtain significant genetic modifications. Thus, we postulated that stabilizing the ZFN protein could enhance ZFN function. However, ZFN stability and the factors that affect it have yet to be investigated. Proteins are in a continual state of flux between synthesis and degradation in a cell. The ubiquitin proteasome pathway is one of the major cellular regulatory mechanisms involved in protein turnover and half-life. UPP plays a key role in eliminating intracellular proteins in eukaryotes, especially misfolded cellular proteins. During ubiquitination, a post-translational modification that targets proteins for degradation by the 26S proteasome, multiple ubiquitin molecules are covalently attached to targeted proteins. This process is catalyzed by a three step cascade mechanism, which involves a ubiquitin activating enzyme, a ubiquitin conjugating enzyme, and a ubiquitin ligase. E1 activates ubiquitin molecules by the formation of an ATP-dependent thiol ester bond between the C-terminus of ubiquitin and the active cysteine site of the E1 enzyme. Activated ubiquitin is transferred to the active cysteine site of the E2 enzyme. Ultimately, E3 catalyzes the transfer of ubiquitin molecules to a lysine residue, ultimately forming polyubiquitin chains on the protein that is  destined for degradation. Finally, ubiquitinated proteins are directed into the 20S core proteolytic KRX-0401 chamber in an ATP-dependent manner for 26S proteasomal degradation. Small chemical molecules, such as synthetic, cell-permeable peptide aldehydes that form covalent adducts with the 20S proteasome and inhibit its peptidase activities, have been developed. Synthetic proteasome inhibitors are peptide aldehydes which are broadly used as inhibitors for both Serine and Cysteine proteases. Several proteasome inhibitors that can enter the cells and block protein degradation pathway have been identified. Among them, the proteasome inhibitor MG132 is the most widely used commercial inhibitor for regulating the UPP. Because ZFN levels are directly proportional to ZFN activity.
destined for degradation. Finally, ubiquitinated proteins are directed into the 20S core proteolytic KRX-0401 chamber in an ATP-dependent manner for 26S proteasomal degradation. Small chemical molecules, such as synthetic, cell-permeable peptide aldehydes that form covalent adducts with the 20S proteasome and inhibit its peptidase activities, have been developed. Synthetic proteasome inhibitors are peptide aldehydes which are broadly used as inhibitors for both Serine and Cysteine proteases. Several proteasome inhibitors that can enter the cells and block protein degradation pathway have been identified. Among them, the proteasome inhibitor MG132 is the most widely used commercial inhibitor for regulating the UPP. Because ZFN levels are directly proportional to ZFN activity.