Other examples include cooperation with Smads, Sp1 or CREB. Additionally, USFs have been shown to complement HIF binding either at neighbouring or identical sites, while collaboration with ETS transcription factors has been proposed to play a role in HIF2a target selectivity. Recent genome-wide approaches relying on experimental and computational identification of HIF binding sites have reported overrepresented transcription factor binding sites in the flanking sequences that might be indicative of transcriptional cooperativity. However, significant differences exist in the overrepresented TFBSs predicted in each study, and the functional significance of these enriched motifs remains unclear. Gene expression profiling indicates that the expression of thousands of genes changes with hypoxia, with vast cell-type differences in the specific genes being regulated. HIF1a ChIP-chip binding Mechlorethamine hydrochloride locations have been reported in cell lines of diverse tissue origin, namely HepG2 hepatocarcinoma cells, MCF-7 breast cancer cells and U87 glioma cells, showing differences in the binding sites identified in each experiment. In previous studies we integrated microarray expression profiling experiments and HIF binding site predictions in a core set of tissue-independent HIF target genes. To further investigate the selectivity of HIF1 binding, in this work we conducted HIF1a ChIP-chip in cervical carcinoma HeLa cells and observed largely non-overlapping binding locations with previous studies. To explore the role of cooperativity in HIF target selection, we integrated HIF1 alpha ChIP-chip binding locations across cell-types with a meta-analysis of gene expression profiles of cells exposed to hypoxia. Computational prediction of enriched transcription factor binding sites in this integrated set suggested several stress-responsive transcription factors as potential HIF1 collaborators. Experimental validation of these predictions in cell-based reporter assays indicates that binding sites for stressresponsive transcription factors other than HIFs, such as CEBPs, contribute to cooperative hypoxic activation of individual targets. To obtain a set of background genomic regions, custom perl scripts were used to exploit the microarray metaanalysis results for the identification of genes harbouring conserved RCGTG motifs but that are unlikely to be modulated by hypoxia. To this end, gene loci that contained conserved RCGTG motifs in their non-coding sequences were first selected. For these genes, each of their probes was examined, and only genes for which all of their associated probes exhibited a mean fold value within 0.25 standard deviations of the global mean in 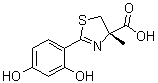 each of the 19 datasets employed in the meta-analysis were considered as not induced by hypoxia. The selected coordinates of conserved RCGTG motifs mapping to these loci were extended as previously described for the set of core HIF binding sites. Genomic regions from this collection were further selected to match the frequency of genomic locations found in the core HBR set. Briefly, Perl scripts were used to annotate core HBRs as promoter, 59UTR, intronic or 39UTR genomic locations and to choose, from the whole collection, a random sample according to the proportions of genomic locations found in the core HBR set. Similarly as with the set of core HIF binding regions, multiple sequence alignments corresponding to the selected control regions were retrieved. The length of flanking non-coding sequences was based on evolutionary conservation, as Chlorhexidine hydrochloride indicated by genomic annotation of PhastCons elements. Statistical assessment of sequence motif enrichment in this set of sequences requires comparison with a background set, the election of this set greatly influencing the results of the analysis.
each of the 19 datasets employed in the meta-analysis were considered as not induced by hypoxia. The selected coordinates of conserved RCGTG motifs mapping to these loci were extended as previously described for the set of core HIF binding sites. Genomic regions from this collection were further selected to match the frequency of genomic locations found in the core HBR set. Briefly, Perl scripts were used to annotate core HBRs as promoter, 59UTR, intronic or 39UTR genomic locations and to choose, from the whole collection, a random sample according to the proportions of genomic locations found in the core HBR set. Similarly as with the set of core HIF binding regions, multiple sequence alignments corresponding to the selected control regions were retrieved. The length of flanking non-coding sequences was based on evolutionary conservation, as Chlorhexidine hydrochloride indicated by genomic annotation of PhastCons elements. Statistical assessment of sequence motif enrichment in this set of sequences requires comparison with a background set, the election of this set greatly influencing the results of the analysis.
Month: June 2019
Both intrauterine and postnatal malnutrition promote the same patterns of Ang II receptor expression
As discussed above, in a similar model of maternal undernutrition, plasma levels of the anorexigenic leptin were increased and levels of the orexigenic prolactin were decreased. In addition to decreasing maternal food intake, this hormone imbalance could have imprinted an impaired dietary behavior on the progeny. Evidence for this concerns elevated plasma levels of leptin in 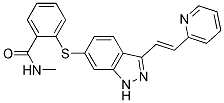 pups from mothers that had been undernourished during lactation, as a result of direct transmission via milk. Acid maceration is not the gold-standard method for estimating the Pimozide number of nephrons. However, reproducibility of counting using Lomitapide Mesylate different kidneys and more than one independent observer, together with comparable control values with data from other laboratories, demonstrated a profound influence of programming in terms of the number of nephrons in adult kidneys. The reduced number of nephrons and the structural glomerular alterations demonstrate that the decreased kidney mass of programmed animals was not a simple relationship to lower body mass. The decreased number of nephrons and decreased capillary area with consequent hypofiltration could be associated with hyperfiltration in remnant healthy nephrons, where the increased intracapillary pressure would contribute to their late and progressive self-destruction with late onset hypertension. The onset of hypertension in older rats supports this view. An increase in the glomerular area would be expected when the number of nephrons is decreased, and compensatory hyperfiltration occurs in some remnant glomeruli. It could be that intense collagen deposition in the cortex of programmed animals affects many glomerular structures, causing a global reduction in the size of the Bowman’s capsule and glomerular capillary tufts, with preservation or an increase in the area of others. It is unexpected that GFR increases by 70% in 60 day-old programmed rats, despite reduced glomerular areas as presented in Fig. 4. However, this could be explained by exacerbated hyperfiltration in the preserved nephrons. The intense proteinuria that accompanies the increase in GRF and UNa reveals important and early damage in the filtration barrier that could evolve into global impairment of renal function in programmed rats. Therefore, kidneys from progeny that were programmed during lactation suffer from early severe morphological, and consequently functional, alterations in glomerular components, adding to the molecular alterations in proximal tubules, as discussed below. The results depicted in Figs. 6, 7, 8, 9 demonstrate that programming during lactation strongly imprints the molecular machinery responsible for the majority of Na + absorption and the fine tuning of this process, leading to augmented recovery of the filtered fluid, which can subsequently contribute to the onset of hypertension. It is interesting that placental undernutrition promotes different modifications from those observed in the present study of young adult rats. This supports the view that depending on the window of development, undernutrition may evoke different signals that affect the same organs in different ways. Up-regulation of AT1 receptors and down-regulation of AT2 receptors could represent the persistence into adulthood of an adaptative response towards impaired RAS functioning in early life. Lower local Ang II production in early life could be compensated by the programming-induced reciprocal alterations in the populations of AT1 and AT2 receptors as demonstrated in Fig. 10, which persisted into adult life. In contrast to the results concerning active Na + transporters.
pups from mothers that had been undernourished during lactation, as a result of direct transmission via milk. Acid maceration is not the gold-standard method for estimating the Pimozide number of nephrons. However, reproducibility of counting using Lomitapide Mesylate different kidneys and more than one independent observer, together with comparable control values with data from other laboratories, demonstrated a profound influence of programming in terms of the number of nephrons in adult kidneys. The reduced number of nephrons and the structural glomerular alterations demonstrate that the decreased kidney mass of programmed animals was not a simple relationship to lower body mass. The decreased number of nephrons and decreased capillary area with consequent hypofiltration could be associated with hyperfiltration in remnant healthy nephrons, where the increased intracapillary pressure would contribute to their late and progressive self-destruction with late onset hypertension. The onset of hypertension in older rats supports this view. An increase in the glomerular area would be expected when the number of nephrons is decreased, and compensatory hyperfiltration occurs in some remnant glomeruli. It could be that intense collagen deposition in the cortex of programmed animals affects many glomerular structures, causing a global reduction in the size of the Bowman’s capsule and glomerular capillary tufts, with preservation or an increase in the area of others. It is unexpected that GFR increases by 70% in 60 day-old programmed rats, despite reduced glomerular areas as presented in Fig. 4. However, this could be explained by exacerbated hyperfiltration in the preserved nephrons. The intense proteinuria that accompanies the increase in GRF and UNa reveals important and early damage in the filtration barrier that could evolve into global impairment of renal function in programmed rats. Therefore, kidneys from progeny that were programmed during lactation suffer from early severe morphological, and consequently functional, alterations in glomerular components, adding to the molecular alterations in proximal tubules, as discussed below. The results depicted in Figs. 6, 7, 8, 9 demonstrate that programming during lactation strongly imprints the molecular machinery responsible for the majority of Na + absorption and the fine tuning of this process, leading to augmented recovery of the filtered fluid, which can subsequently contribute to the onset of hypertension. It is interesting that placental undernutrition promotes different modifications from those observed in the present study of young adult rats. This supports the view that depending on the window of development, undernutrition may evoke different signals that affect the same organs in different ways. Up-regulation of AT1 receptors and down-regulation of AT2 receptors could represent the persistence into adulthood of an adaptative response towards impaired RAS functioning in early life. Lower local Ang II production in early life could be compensated by the programming-induced reciprocal alterations in the populations of AT1 and AT2 receptors as demonstrated in Fig. 10, which persisted into adult life. In contrast to the results concerning active Na + transporters.
Parameters and metrics for working specifically with EST transcriptomic sequences produced by 454 sequencers
It was our last choice of the tested assemblers, according to evaluation metrics used. We also verified that some software works better with different clustering measures, such as the Celera assembler, which more effectively maps all the assembled reads into clusters. Most researchers would agree that the problem of sequence assembly and consensus production is highly complex and has not yet been solved, although most papers in the field simply choose a software program and proceed with assembly without further evaluations. Because sequence clustering is dependent on the informational content of the original reads, we recommend that authors test the appropriateness of the software for their own data before commencing the Lomitapide Mesylate annotation and analysis of unigenes. Based on the number of unigenes found here, we suspect that we have sequenced most of the coding sequences expressed by Actinopus spp. and G. cancriformis in their spinning glands. The number of unigenes in the Unigene database for the tick I. scapularis suggests that a number of unigenes in excess of the 25,663 found for G. cancriformis may be the rule for the Chelicerata clade. This excess of unigenes cannot be explained simply by the small size of the NGS-generated unigenes, although many unigenes probably represent either 39 or the 59 portions of genes. Our inability to identify most of the G. cancriformis unigenes by BLAST analysis indicates a lack of interest in Chelicerata clade genomics. We tried to fix this knowledge gap in the present publication by identifying more than 30,000 new genes for this clade and making them available to the research community in the SRA database. The amazing genetic Butenafine hydrochloride repertoire of spiders with their highly specialized tissues, such as the spinning and venom glands, will help researchers make biotechnological breakthroughs in the coming years, as has been recently discussed in the specialized literature. Remarkably, we found the metalloproteases from the astacin family to be extremely significant in the G. cancriformis spinning gland transcriptome. Genes from this family, which still have unknown functions, corresponded to 1.8% of all transcripts sequenced in the spinning glands, a number approximately 3.6 times larger than the number of expressed spidroin genes. Moreover, among the gene families that have undergone major amplifications since the ancestral split between the Mygalomorphae and Araneomorphae clades, we found that this family of metal-dependent proteinases has been amplified 10 times in the genome of the Araneomorphae/Orbiculariae 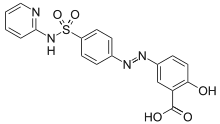 clade. The observed proteolytic effects of metalloproteinases in the venom of some Araneomorphae spiders are probably unrelated to their high expression in the spinning glands. Moreover, until recently, this subfamily of the meprin metalloproteases was believed to exist only in vertebrates, although even more recently, it was reported in the horseshoe crab Limulus polyphemus, an organism from the subphylum Chelicerata. Our work, therefore, corroborates the evidence that this gene family is present in the Arthropoda subphylum studied here. Moreover, KOG data from 2003 have classified a number of these proteins in the genomes of invertebrates such as D. melanogaster and Caenorhabditis elegans. Meprins consist of a single, membrane-anchored member of the astacin family and have been shown to be implicated in a number of complex cellular processes in higher eukaryotes, such as cell migration, immune reactions, and tissue differentiation. A clue to their function may come from some works reporting their relationship to human collagen fibers.
clade. The observed proteolytic effects of metalloproteinases in the venom of some Araneomorphae spiders are probably unrelated to their high expression in the spinning glands. Moreover, until recently, this subfamily of the meprin metalloproteases was believed to exist only in vertebrates, although even more recently, it was reported in the horseshoe crab Limulus polyphemus, an organism from the subphylum Chelicerata. Our work, therefore, corroborates the evidence that this gene family is present in the Arthropoda subphylum studied here. Moreover, KOG data from 2003 have classified a number of these proteins in the genomes of invertebrates such as D. melanogaster and Caenorhabditis elegans. Meprins consist of a single, membrane-anchored member of the astacin family and have been shown to be implicated in a number of complex cellular processes in higher eukaryotes, such as cell migration, immune reactions, and tissue differentiation. A clue to their function may come from some works reporting their relationship to human collagen fibers.
Phosphoserine and phosphothreonine containing peptides may produce low scores due to fragmentation
Predicted to phosphorylate Bcl2-associated athanogene 3, Maf1 and peroxisome biogenesis factor 1. In recent years, several groups have performed comprehensive tissue phosphoproteome analyses. The animal tissue most often used for phosphoproteome analysis has been liver. The first such study, performed by Jin et al., utilized iron IMAC for the enrichment of phosphorylated peptides and conventional linear ion-trap mass spectrometry for the phosphopeptide analysis. These investigators identified 26 nonredundant phosphorylation sites. A subsequent study utilized high capacity iron IMAC and a higher mass accuracy MS Q-TOF instrument to identify 339 non-redundant phosphorylation sites from over 200 proteins. The analyses performed by Villen et al. in 2007 was a breakthrough study. These investigators identified 5,635 nonredundant phosphorylation sites from 2,149 proteins, signifying the first in-depth global analysis of phosphopeptides from liver. Notably, these investigators used more selective iron IMAC beads and MS instruments of higher mass accuracy as compared to previous studies. However, it is likely that the SCX pre-fractionation of tryptic peptides was a critical factor accounting for the increase in phosphopeptide identification. Lack of reproducibility has been an issue with large scale MSbased phosphoproteomic profiling. Moser and White tested the reproducibility of their methodology by performing three replicate analyses of the same rat liver homogenate. They observed that 56�C63% of the peptides from each analysis were observed in all three analyses. Another issue is method of analysis. Alcolea et al. found that analyzing the same phosphopeptide enriched murine NIH/3T3 fibroblast lysate by two different LC-MS/MS Gomisin-D platforms based on Q-TOF and LTQ-Orbitrap mass spectrometers led to identification 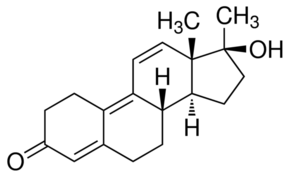 of partially overlapping, but also distinct, phosphoproteome profiles. The QTOF based platform resulted in 1,485 non-redundant phosphopeptide identifications, whereas the LTQ-Orbitrap based platform identified 4,308 non-redundant phosphopeptides. Only 1,077 of the total population of phosphopeptides were detected by both platforms. Analyzing duplicate samples by LC-MS/MS on the LTQ-Orbitrap platform showed that,70% of the identified phosphopeptides were identical. In a study comparing the peptides identified using two workflows, TiO2-SCX and SCX-TiO2, the overlap was 58 and 51% for the two methods, respectively. A similar overlap of 60% was observed, when they performed replicate LC-MS/MS analyses of the same TiO2-SCX sample by an LTQ-Orbitrap. These previous studies profiling the liver phosphoproteome have not been aimed at characterizing signaling events in a physiological context. Accomplishing this required LOUREIRIN-B improved sensitivity and accuracy, and a demonstration of reproducibility. We found that peptide abundance affects reproducibility, but that reproducibility could be enhanced by the performance of technical replicates. There are several indications that our methods were sufficient to detect mTORC1-mediated protein phosphorylation. These included the identification of known mTORC1 targets, the significant enrichment for mTOR signaling pathway constituents as indicated by pathway analysis, and the kinase prediction results. In their seminal studies, Hunter and Sefton reported the relative abundances of pSer, pThr, and pTyr to be 90%, 10%, and 0.05%,respectively. The order of magnitude higher tyrosine phosphorylation frequency in our results may be a function of the tendency for pTyr-containing peptides to yield better quality MS/MS spectra with collision-induced dissociation fragmentation.
of partially overlapping, but also distinct, phosphoproteome profiles. The QTOF based platform resulted in 1,485 non-redundant phosphopeptide identifications, whereas the LTQ-Orbitrap based platform identified 4,308 non-redundant phosphopeptides. Only 1,077 of the total population of phosphopeptides were detected by both platforms. Analyzing duplicate samples by LC-MS/MS on the LTQ-Orbitrap platform showed that,70% of the identified phosphopeptides were identical. In a study comparing the peptides identified using two workflows, TiO2-SCX and SCX-TiO2, the overlap was 58 and 51% for the two methods, respectively. A similar overlap of 60% was observed, when they performed replicate LC-MS/MS analyses of the same TiO2-SCX sample by an LTQ-Orbitrap. These previous studies profiling the liver phosphoproteome have not been aimed at characterizing signaling events in a physiological context. Accomplishing this required LOUREIRIN-B improved sensitivity and accuracy, and a demonstration of reproducibility. We found that peptide abundance affects reproducibility, but that reproducibility could be enhanced by the performance of technical replicates. There are several indications that our methods were sufficient to detect mTORC1-mediated protein phosphorylation. These included the identification of known mTORC1 targets, the significant enrichment for mTOR signaling pathway constituents as indicated by pathway analysis, and the kinase prediction results. In their seminal studies, Hunter and Sefton reported the relative abundances of pSer, pThr, and pTyr to be 90%, 10%, and 0.05%,respectively. The order of magnitude higher tyrosine phosphorylation frequency in our results may be a function of the tendency for pTyr-containing peptides to yield better quality MS/MS spectra with collision-induced dissociation fragmentation.
Since DENV interacts with heparan sulfate syndecan-3 may be a possible receptor on DC
It has been hypothesized that DENV needs DC-SIGN for attachment and enhancing infection of DC in cis and needs MR for internalization. In fact, cells expressing mutant DC-SIGN, lacking the internalization domain, are still susceptible for DENV infection because DC-SIGN can capture the pathogen. Interaction between DENV and DC-SIGN or MR is abrogated by deglycosylation of the DENV envelope and by EDTA or mannan, indicating that the interaction is carbohydrate dependent. DC-SIGN and MR have respectively 1 and 8 carbohydrate recognition domains responsible for pathogen recognition. Other pathogens recognized by DCSIGN and MR are HIV, HCV and human cytomegalovirus. These interactions are carbohydrate-dependent and are inhibited by various CBAs. In a previous report, we were the first to demonstrate the antiviral activity of the CBAs against DENV-2 in Raji/DC-SIGN cells and IL-4-treated monocytes. In the Cinoxacin present study, we studied the antiviral activity of these CBAs on the four DENV serotypes in primary MDDC, the most important target cells for DENV. A number of these CBAs proved about 100-fold  more effective in inhibiting DENV infection in primary MDDC compared to the transfected Raji/DC-SIGN cell line. We also demonstrated that the mannose binding lectin HHA prevents DENV-2 binding to the host cell and acts less efficiently in the postbinding stage. HHA interacts with DENV and not with DC-SIGN on the target cell. The potency of HHA to inhibit attachment of DENV to Raji/DC-SIGN cells is Ginsenoside-Ro comparable to its inhibitory activity of the capture of HIV and HCV to Raji/DCSIGN + cells. CBAs could thus be considered as unique prophylactic agents. However, plant lectins are not orally bioavailable, sensitive for proteolytic cleavage and expensive to produce, they provide novel insights into the entry mechanism of DENV in human primary cells. The search for non-peptidic small molecules with CBA-like activity is therefore warranted. PRM-S, a derivate of the antibiotic PRM-A, acts as a CBA in terms of glycan recognition and exerts antiviral activity against HIV and SIV. The compound has high solubility and a high barrier for HIV resistance development. In MDDC, we observed a dose-dependent antiviral activity of PRM-S against DENV-2, comparable to the antiviral activity against HIV. In contrast, PRM-S exerted only weak antiviral activity in Raji/DCSIGN + cells. Accordingly, the antiviral potency of the other CBAs, HHA, GNA and UDA was higher in primary MDDC than in Raji/DC-SIGN + cells as well. This may be due to several celldependent specificities. First, Raji/DC-SIGN + cells are more susceptible for DENV infection compared to MDDC although the DC-SIGN expression level is comparable in the two cell-types. Although MDDC cultures contain a low amount of T-cells and B-cells, these cell types are not susceptible for DENV infection. Second, the entry process of DENV in Raji/DC-SIGN + cells and in MDDC is fundamentally different. In Raji/DCSIGN + cells, the entry process is mainly dependent on DC-SIGN. This is in contrast to MDDC, where unidentified cofactors for infection or DC-SIGN-independent entry pathways of the virus may be present. Although, whatever entry pathway in MDDC is employed by the virus, DENV infection is efficiently inhibited by CBAs. This indicates that the DENV entry in human cells is carbohydrate-dependent and that CBAs also inhibit DC-SIGNindependent entry pathways in MDDC. Consequently, the observed antiviral activity of the CBAs in human primary MDDC may be considered more relevant than in artificial constructed cell lines such as the Raji/DC-SIGN + cell line.
more effective in inhibiting DENV infection in primary MDDC compared to the transfected Raji/DC-SIGN cell line. We also demonstrated that the mannose binding lectin HHA prevents DENV-2 binding to the host cell and acts less efficiently in the postbinding stage. HHA interacts with DENV and not with DC-SIGN on the target cell. The potency of HHA to inhibit attachment of DENV to Raji/DC-SIGN cells is Ginsenoside-Ro comparable to its inhibitory activity of the capture of HIV and HCV to Raji/DCSIGN + cells. CBAs could thus be considered as unique prophylactic agents. However, plant lectins are not orally bioavailable, sensitive for proteolytic cleavage and expensive to produce, they provide novel insights into the entry mechanism of DENV in human primary cells. The search for non-peptidic small molecules with CBA-like activity is therefore warranted. PRM-S, a derivate of the antibiotic PRM-A, acts as a CBA in terms of glycan recognition and exerts antiviral activity against HIV and SIV. The compound has high solubility and a high barrier for HIV resistance development. In MDDC, we observed a dose-dependent antiviral activity of PRM-S against DENV-2, comparable to the antiviral activity against HIV. In contrast, PRM-S exerted only weak antiviral activity in Raji/DCSIGN + cells. Accordingly, the antiviral potency of the other CBAs, HHA, GNA and UDA was higher in primary MDDC than in Raji/DC-SIGN + cells as well. This may be due to several celldependent specificities. First, Raji/DC-SIGN + cells are more susceptible for DENV infection compared to MDDC although the DC-SIGN expression level is comparable in the two cell-types. Although MDDC cultures contain a low amount of T-cells and B-cells, these cell types are not susceptible for DENV infection. Second, the entry process of DENV in Raji/DC-SIGN + cells and in MDDC is fundamentally different. In Raji/DCSIGN + cells, the entry process is mainly dependent on DC-SIGN. This is in contrast to MDDC, where unidentified cofactors for infection or DC-SIGN-independent entry pathways of the virus may be present. Although, whatever entry pathway in MDDC is employed by the virus, DENV infection is efficiently inhibited by CBAs. This indicates that the DENV entry in human cells is carbohydrate-dependent and that CBAs also inhibit DC-SIGNindependent entry pathways in MDDC. Consequently, the observed antiviral activity of the CBAs in human primary MDDC may be considered more relevant than in artificial constructed cell lines such as the Raji/DC-SIGN + cell line.