Although use of 2A technology has previously been reported in plants, Samalova and coworkers have questioned its use in plant systems, demonstrating that further development and characterization of the 2A system is necessary. In this study, we wanted to improve the 2A technology for use in plant cells as a tool to study basic cellular processes, such as protein trafficking mechanisms. After successful optimization of 2A cleavage efficiency, mutant forms of small GTP-binding proteins could be expressed and used to interfere with trafficking of CAH1 at the endomembrane system. The results presented in this work highlight the 2A technology as a valuable tool for effective and stoichiometric co-expression of marker and effector molecules in plant systems. In addition, our study demonstrates that 2A mediated transient co-expression of fluorescent markers combined with fluorescent activated cell sorting can be used to obtain homogeneous mutant protoplast populations in very short time. Our data shows that similar to RABD2a, dominant mutant versions of SAR1 and ARF1 originating from 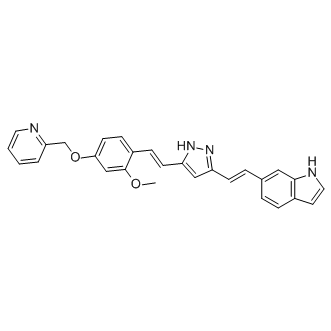 the GSCFP-2A construct were functional and gave a phenotypic effect on the Golgi-localized marker protein. The effects were consistent to those observed in previous studies. Our findings clearly demonstrate the value of a stoichiometric co-expression system that can easily be applied to live-cell imaging and confocal microscopy studies. These results prompted us to further extend and test the versatility of the 2A system in combination with other experimental techniques, such as biochemical assays and fluorescence-activated cell sorting. To Mechlorethamine hydrochloride continue our study of the mechanisms involved in transport of CAH1 between the ER and the Golgi, we explored the requirement for specific GTPases involved in vesicle trafficking. Although BFA arrested CAH1-GFP in aggregate-like structures, it is known to have different effects in different species, and even in different tissues within a species. In addition, high BFA concentrations likely induce secondary effects, alterations that can be reduced using genetic tools. The potential involvement of three individual GTPases was tested in this initial study; RABD2a, SAR1, and ARF1, all of them important for vesicle formation or docking at the ER-Golgi interface. Specific single point mutations in these GTPases can generate arrested and non-functional enzymes. RABD2a is a small GTPase involved in targeting and fusion of ER-derived COPII vesicles at the Golgi surface. Dominant negative variants where an N121I substitution was introduced in the GTP binding motif ), were shown to inhibit trafficking of secreted and Golgi targeted proteins out from the ER. On the contrary, SAR1 and ARF1 are directly involved in the formation of COPII and COPI vesicles, respectively. Two mutant Chlorhexidine hydrochloride isoforms of ARF1 were tested: ARF1 and ARF1, both affecting ER-to-Golgi trafficking and relocating Golgi markers to the ER. The ARF1 mutant shows reduced GTPase activity, therefore acting as a constitutively activated mutant, interfering with sorting of membrane proteins into Golgiderived COPI vesicles. The ARF1 mutant instead has low affinity for GTP, acting as a dominant-negative mutant that blocks formation of COPI vesicles. We induced ARF1 expression under the control of a heat-shock promoter in fourweeks-old Arabidopsis plants. Unfortunately, ARF1 expression in leaf tissue was very low, which in combination with the time needed for obtaining good chloroplast preparations, prompted us to seek a different experimental system.
the GSCFP-2A construct were functional and gave a phenotypic effect on the Golgi-localized marker protein. The effects were consistent to those observed in previous studies. Our findings clearly demonstrate the value of a stoichiometric co-expression system that can easily be applied to live-cell imaging and confocal microscopy studies. These results prompted us to further extend and test the versatility of the 2A system in combination with other experimental techniques, such as biochemical assays and fluorescence-activated cell sorting. To Mechlorethamine hydrochloride continue our study of the mechanisms involved in transport of CAH1 between the ER and the Golgi, we explored the requirement for specific GTPases involved in vesicle trafficking. Although BFA arrested CAH1-GFP in aggregate-like structures, it is known to have different effects in different species, and even in different tissues within a species. In addition, high BFA concentrations likely induce secondary effects, alterations that can be reduced using genetic tools. The potential involvement of three individual GTPases was tested in this initial study; RABD2a, SAR1, and ARF1, all of them important for vesicle formation or docking at the ER-Golgi interface. Specific single point mutations in these GTPases can generate arrested and non-functional enzymes. RABD2a is a small GTPase involved in targeting and fusion of ER-derived COPII vesicles at the Golgi surface. Dominant negative variants where an N121I substitution was introduced in the GTP binding motif ), were shown to inhibit trafficking of secreted and Golgi targeted proteins out from the ER. On the contrary, SAR1 and ARF1 are directly involved in the formation of COPII and COPI vesicles, respectively. Two mutant Chlorhexidine hydrochloride isoforms of ARF1 were tested: ARF1 and ARF1, both affecting ER-to-Golgi trafficking and relocating Golgi markers to the ER. The ARF1 mutant shows reduced GTPase activity, therefore acting as a constitutively activated mutant, interfering with sorting of membrane proteins into Golgiderived COPI vesicles. The ARF1 mutant instead has low affinity for GTP, acting as a dominant-negative mutant that blocks formation of COPI vesicles. We induced ARF1 expression under the control of a heat-shock promoter in fourweeks-old Arabidopsis plants. Unfortunately, ARF1 expression in leaf tissue was very low, which in combination with the time needed for obtaining good chloroplast preparations, prompted us to seek a different experimental system.
Month: June 2019
Reorganization effects mainly on cell adhesion proteins because they are directly connected to cytoskeleton
For example, the reduced cytoskeletal reorganization will weaken the cytoskeletal engagement of integrins in tetraspanin-enriched microdomains and subsequently lead to the local attenuation of integrin signaling or even inactivation of integrins. We extrapolate that both imbalance of Rho GTPases and inactivation of integrins result in the aberrancy in cellular morphology and diminishment in cell motility. Consistent with the current understanding of the roles of Rac in cell morphology and movement, the suppressed lamellipodia and cell movement upon KAI1/CD2 expression correlated well with the diminished Rac1 activity in Du145 cells. In some cell types, Rac1 activation negatively regulates RhoA activity by generating reactive oxygen species and subsequently activating p190RhoGAP at the plasma membrane. The delicate balance between the antagonistic activities of Rac1 and RhoA is crucial for proper cell movement and also specifies cell morphology. In addition, KAI1/CD82 was reported to inhibit the activity of Src kinases, which activates p190RhoGAP by tyrosine phosphorylation, and the lower Src activity ultimately leads to the RhoA activation. However, the constitutive RhoA activity appears to be unaltered upon KAI1/CD82 expression. Although HGF markedly enhanced RhoA activity, KAI1/CD82 apparently could block such stimulation, reflected by the defect in retraction and the formation of fewer stress fibers in KAI1/Mepiroxol CD82-expressing cells. It remains unclear that, in Du145KAI1/CD82 cells, RhoA still exhibits a similar level of constitutive activity, although it cannot reach the plasma membrane. As a key effector of both Rac and Rho, cofilin plays an important role in membrane ruffling or lamellipodia formation. Driven by activated Rac or PIP2, cofilin is translocated to or enriched in the cell periphery where it interacts with actin cytoskeleton, generates more barbed ends, and promotes actin cortical meshwork formation and consequently lamellipodia formation. Translocation of cofilin to the plasma membrane is considered to be an indicator of cofilin activation. Interestingly, the subcellular localizations of total and inactivated cofilin in Du145-Mock and -KAI1/CD82 cells displayed distribution patterns similar to the ones in migrating and nonmigrating fibroblast cells, respectively. Upon KAI1/ CD82 expression, cofilin was no longer enriched at the cell periphery, although it could translocate to the peripheral cytoplasm. This observation strongly suggests that KAI1/CD82 expression blocks the translocation and therefore activation of cofilin, underlining a mechanism by which KAI1/CD82 impairs lamellipodia formation. If so, 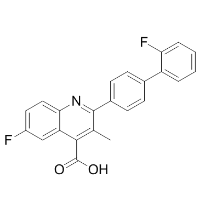 one would expect more phosphorylated or inactive cofilin in Du145-KAI1/CD82 cells. However, the level of phosphorylated cofilin proteins remained unchanged upon KAI1/CD82 expression. Possibly, cofilin in Du145-KAI1/ CD82 cells can still undergo de4-(Benzyloxy)phenol phosphorylation during translocation to the peripheral cytoplasm but cannot become enriched at the cell periphery due to aberrant plasma membrane in Du145KAI1/CD82 cells. When KAI1/CD82 is present, HGF cannot upregulate the activities of ROCK and its direct upstream activator RhoA. Besides agreeing with earlier observations that KAI1/CD82 inhibits HGF/c-Met signaling, these findings have further demonstrated that CD82 intercepts the HGF/c-Met signaling leading to the cellular retraction. Because KAI1/CD82 can inhibit the retraction process even without HGF stimulation, we predict that the local, constitutive ROCK activity at the retraction areas in KAI1/CD82-expressing cells.
one would expect more phosphorylated or inactive cofilin in Du145-KAI1/CD82 cells. However, the level of phosphorylated cofilin proteins remained unchanged upon KAI1/CD82 expression. Possibly, cofilin in Du145-KAI1/ CD82 cells can still undergo de4-(Benzyloxy)phenol phosphorylation during translocation to the peripheral cytoplasm but cannot become enriched at the cell periphery due to aberrant plasma membrane in Du145KAI1/CD82 cells. When KAI1/CD82 is present, HGF cannot upregulate the activities of ROCK and its direct upstream activator RhoA. Besides agreeing with earlier observations that KAI1/CD82 inhibits HGF/c-Met signaling, these findings have further demonstrated that CD82 intercepts the HGF/c-Met signaling leading to the cellular retraction. Because KAI1/CD82 can inhibit the retraction process even without HGF stimulation, we predict that the local, constitutive ROCK activity at the retraction areas in KAI1/CD82-expressing cells.
These cells retain the characteristics of proximal renal tubular epithelium and have been used successfully
For example, miR-127 has been shown to participate in cancer development, miR-145 has been shown to control c-Myc expression through p53, miR-199a Mechlorethamine hydrochloride regulates MET protooncogene and affects NF-KB expression, miR-379 affects brain neuronal development, miR-451 affects erythroid differentiation, miR-126 affects angiogenic signaling and controls blood vessel development, miR-143 regulates ERK5 signaling and targets KRAS gene, miR-298 regulates CYPA3 expression and miR-486 regulates kinase activity and tumor progression. Also, miR-671 and miR-700 are involved cancer growth and development. miR-669 is involved in c-Myc expression through p53, miR-500 regulates MET protooncogenes and affects NF-kB, miR-466 is involved in mammary tumor development, miR-466c is involved in tumor growth, miR-449a regulates breast cancer development and inhibits cell proliferation and miR-Let7b plays a role in myeloid leukemia. Together, such data suggested that TCDD affects a large number of miRs that may be directly or indirectly involved in tumor induction and promotion. The precise role of such miRs in TCDD-induced tumorigenesis and toxicity in vivo can be better addressed by using mice deficient in such miRs. In summary, we demonstrate for the first time that exposure to environmental toxicants such as 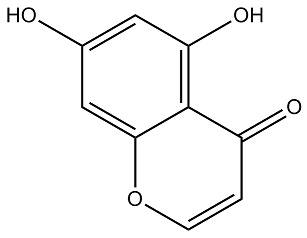 TCDD during pregnancy can have a significant effect on the miR profile of fetal thymus and thereby influence the regulation of a large number of genes that may affect the development of the immune system. Identification of miRs as targets for TCDD-induced modulation of gene expression offers insights into novel pathways to further understand the mechanisms of toxicity. Oxalate is a metabolic end product that is freely filtered at the glomerulus, undergoes bi-directional transport in the renal tubules, and is excreted primarily by the kidney. The most common pathological condition involving oxalate is the formation of calcium oxalate stones in the kidney. While very high levels of urinary oxalate are observed only in subjects with primary hyperoxaluria, a majority of idiopathic kidney stone patients only show a mild elevation in urinary oxalate In addition several other conditions associated with oxalate deposits are: renal cysts in acquired renal cystic disease, proliferating cells in the kidney, hyperplasic thyroid glands, and benign neoplasm of the breast. These considerations suggest that the pathological deposition of calcium oxalate is more complex than a simple physical precipitation of calcium oxalate crystals. In 1994, we were the first group to note that oxalate renal cell interactions involved alterations in gene expression. Over the past two decades, studies have demonstrated that oxalate interactions with renal epithelial cells result in a program of events, including changes in gene expression and cell dysfunction, consistent with cellular stress. Studies from our laboratory demonstrated that oxalate induced changes in renal cells are inhibited by inhibitors of transcription and translation, indicating that the cellular response to oxalate toxicity is dependent on new gene expression and protein synthesis. Moreover, cells of the renal Tulathromycin B tubular epithelium are exposed to an environment with variable and elevated concentrations of the oxalate and must be able to adapt to oxalate stress. Indeed we have shown that many signal transduction pathways, including p38 MAPK and JNK, are activated in renal epithelial cells in response to oxalate and COM crystals. However, the genetic response of renal epithelial cells to oxalate exposure remains ambiguous. HK-2 cells are a line of human proximal tubular epithelial cells immortalized by using the E6/E7 genes of human papilloma virus.
TCDD during pregnancy can have a significant effect on the miR profile of fetal thymus and thereby influence the regulation of a large number of genes that may affect the development of the immune system. Identification of miRs as targets for TCDD-induced modulation of gene expression offers insights into novel pathways to further understand the mechanisms of toxicity. Oxalate is a metabolic end product that is freely filtered at the glomerulus, undergoes bi-directional transport in the renal tubules, and is excreted primarily by the kidney. The most common pathological condition involving oxalate is the formation of calcium oxalate stones in the kidney. While very high levels of urinary oxalate are observed only in subjects with primary hyperoxaluria, a majority of idiopathic kidney stone patients only show a mild elevation in urinary oxalate In addition several other conditions associated with oxalate deposits are: renal cysts in acquired renal cystic disease, proliferating cells in the kidney, hyperplasic thyroid glands, and benign neoplasm of the breast. These considerations suggest that the pathological deposition of calcium oxalate is more complex than a simple physical precipitation of calcium oxalate crystals. In 1994, we were the first group to note that oxalate renal cell interactions involved alterations in gene expression. Over the past two decades, studies have demonstrated that oxalate interactions with renal epithelial cells result in a program of events, including changes in gene expression and cell dysfunction, consistent with cellular stress. Studies from our laboratory demonstrated that oxalate induced changes in renal cells are inhibited by inhibitors of transcription and translation, indicating that the cellular response to oxalate toxicity is dependent on new gene expression and protein synthesis. Moreover, cells of the renal Tulathromycin B tubular epithelium are exposed to an environment with variable and elevated concentrations of the oxalate and must be able to adapt to oxalate stress. Indeed we have shown that many signal transduction pathways, including p38 MAPK and JNK, are activated in renal epithelial cells in response to oxalate and COM crystals. However, the genetic response of renal epithelial cells to oxalate exposure remains ambiguous. HK-2 cells are a line of human proximal tubular epithelial cells immortalized by using the E6/E7 genes of human papilloma virus.
The group composed by tumor samples seems to present two subgroups according to the protein expression profiling
Our proteomic analysis revealed that several enzymes of the citrate cycle and of oxidative phosphorylation were down-regulated in GC cells. These data show possible alterations in mitochondrion Cinoxacin function and a shift in energy production in the present GC cells, suggesting the Warburg effect. Proliferating tumor cells reprogram their metabolic pathways to generate energy and, thus, support the rapid cell division under stressful metabolic conditions that are characteristic of the abnormal tumor microenvironment. Even under normal oxygen concentrations, tumor cells shift from ATP generation through oxidative phosphorylation to ATP generation through glycolysis, converting most incoming glucose to lactate. It has been proposed that highly active glycolysis provides a biosynthetic advantage for tumor cells. Glycolysis provides enough metabolic intermediates by avoiding the oxidation of glucose, which is essential for the synthesis of macromolecules, such as lipids, proteins, and nucleic acids, during cell division. The lactate dehydrogenase and pyruvate dehydrogenase complexes control the metabolism of pyruvic acids, transforming to either lactic acids or acetyl-CoA 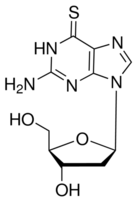 then entering the citrate cycle. The down-regulation of subunits of the LDH and PDH complexes suggests a reduction in pyruvate flux into the citrate cycle and a decrease in the rate of oxidative phosphorylation and oxygen consumption, reinforcing the glycolytic phenotype. Other down-regulated proteins, such as LIPF and GOT1, highlight the activation of other metabolic pathways with the impairment of the citrate cycle and oxidative phosphorylation. Several metabolic alterations that we observed in noncardia GC were also described by Cai et al. in a cardia GC proteomic study, suggesting that these metabolic alterations are not specific to a GC subtype based on tumor location. By the PANTHER system, the most significantly enriched pathway is the p53 pathway observed in the comparison between tumor and control samples. Additionally to its function in the DNA damage response and apoptosis, p53 is also a regulator of cell metabolism. p53 promotes oxidative phosphorylation and also inhibits the glycolytic pathway by up-regulating the expression of TP53-induced glycolysis and the apoptosis regulator. Therefore, the loss of p53 contributes to the acquisition of glycolytic phenotype. The loss of TP53 locus is a common finding in GC of individuals from Northern Brazil. Moreover, 20% of the GC analyzed by 2-DE presented p53 immunoreactivity. The p53 immunoreactivity usually depends on accumulation of mutated proteins in the cell, which leads to a longer half-life. The top networks of molecular interactions and functions were also identified using the IPA software. We showed several differently regulated proteins involved in cellular assembly and organization, and in inflammatory processes. The cellular assembly and organization was the principally enriched network observed in the comparison between controls and tumors with lymph node metastasis. Therefore, our data reveal that the molecules of the described subnetwork are important to the process of metastasis in noncardia Lomitapide Mesylate gastric carcinogenesis. Previous studies have demonstrated that GC is strongly linked to chronic inflammation, and that infection with H. pylori may trigger the chronic inflammation that can lead to malignancy. However, the exact mechanism of this process is still not known. The identified proteins add new pieces to this process in gastric carcinogenesis. The unsupervised hierarchical clustering of the differentially expressed proteins revealed that the tumors and control samples do not form two distinct separate clusters. Although, hierarchical clustering revealed one group composed by only controls, the other group presented all tumor samples and two misclassified control samples.
then entering the citrate cycle. The down-regulation of subunits of the LDH and PDH complexes suggests a reduction in pyruvate flux into the citrate cycle and a decrease in the rate of oxidative phosphorylation and oxygen consumption, reinforcing the glycolytic phenotype. Other down-regulated proteins, such as LIPF and GOT1, highlight the activation of other metabolic pathways with the impairment of the citrate cycle and oxidative phosphorylation. Several metabolic alterations that we observed in noncardia GC were also described by Cai et al. in a cardia GC proteomic study, suggesting that these metabolic alterations are not specific to a GC subtype based on tumor location. By the PANTHER system, the most significantly enriched pathway is the p53 pathway observed in the comparison between tumor and control samples. Additionally to its function in the DNA damage response and apoptosis, p53 is also a regulator of cell metabolism. p53 promotes oxidative phosphorylation and also inhibits the glycolytic pathway by up-regulating the expression of TP53-induced glycolysis and the apoptosis regulator. Therefore, the loss of p53 contributes to the acquisition of glycolytic phenotype. The loss of TP53 locus is a common finding in GC of individuals from Northern Brazil. Moreover, 20% of the GC analyzed by 2-DE presented p53 immunoreactivity. The p53 immunoreactivity usually depends on accumulation of mutated proteins in the cell, which leads to a longer half-life. The top networks of molecular interactions and functions were also identified using the IPA software. We showed several differently regulated proteins involved in cellular assembly and organization, and in inflammatory processes. The cellular assembly and organization was the principally enriched network observed in the comparison between controls and tumors with lymph node metastasis. Therefore, our data reveal that the molecules of the described subnetwork are important to the process of metastasis in noncardia Lomitapide Mesylate gastric carcinogenesis. Previous studies have demonstrated that GC is strongly linked to chronic inflammation, and that infection with H. pylori may trigger the chronic inflammation that can lead to malignancy. However, the exact mechanism of this process is still not known. The identified proteins add new pieces to this process in gastric carcinogenesis. The unsupervised hierarchical clustering of the differentially expressed proteins revealed that the tumors and control samples do not form two distinct separate clusters. Although, hierarchical clustering revealed one group composed by only controls, the other group presented all tumor samples and two misclassified control samples.
Both GATA2 and AP1 binding sites are necessary for epithelial induction of ET-1 under hypoxia
By the evolutionarily conserved Hypoxia Inducible Factor family of basic helix-loop-helix transcription factors. HIFs are heterodimers of a beta subunit, and an alpha subunit. While ARNT levels are not sensitive to oxygen, both HIFa stability and its transcriptional activity are regulated by oxygen-dependent hydroxylation. Under oxygen restriction, HIFa subunits escape  proteasomal degradation, heterodimerize with HIFb subunits and translocate to the cell nucleus, where they bind the RCGTG consensus sequence within regulatory regions of target genes, leading to their transcriptional activation in hypoxia. Mammals present three isoforms of HIFa that differ in their tissue distribution, HIF1a being the more ubiquitous and best characterized. A large number of studies focusing on single genes have identified individual HIF targets that, collectively, account for the functional responses to hypoxia, mainly metabolic adaptation and induction of angiogenesis. More recently, works employing HIF1a and HIF2a chromatin immunoprecipitation coupled to genomic microarrays or high-throughput sequencing have addressed the genome-wide identification of HIF binding locations, thereby improving the existing knowledge on the HIF-modulated transcriptome and largely confirming the RCGTG HIF binding consensus. Additionally, these studies have provided important insights into the global properties of HIF1 binding and transactivation. First, these works reported a significant association between the presence of a HIF binding site and hypoxic induction of the neighboring genes. The same trend was not found for genes repressed by hypoxia, Folinic acid calcium salt pentahydrate suggesting that hypoxia-mediated repression is largely indirect or HIF independent. Furthermore, they have clearly shown that only a small subset of about a hundred of all RCGTG-containing genes is robustly regulated by hypoxia. Hence, and in agreement with work on other transcription factors, HIFs bind a small proportion of potential binding sites, albeit the basis of their binding and target selectivity are incompletely understood. Understanding the mechanisms that explain HIFs transactivation selectivity is of paramount importance to expand our knowledge on transcriptional regulation and to improve the sensitivity and specificity of genome-wide efforts to characterize the HIF transcriptional response. DNA accessibility of transcription factor binding sites can clearly contribute to binding selectivity. For HIFs, recent evidence includes enhanced HIF1 and HIF2 binding to normoxic DNAse hypersensitivity sites and enrichment of HIF1 binding in the proximity of genes with a “permissive” transcriptional state in normoxia, as evidenced by significant basal expression. Additionally, DNA methylation has been also shown to modulate HIF1 binding, as originally demonstrated for the 39 enhancer of the Lomitapide Mesylate erythropoietin gene. A further mechanism that can impact target selectivity is direct or indirect cooperativity between transcription factors. Models of direct cooperativity have been mainly derived from developmental enhancers, and include the strict enhanceosome model, where cooperative occupancy occurs through extensive protein-protein interactions between TFs or common cofactors, and the more flexible billboard model, which suggests that enhancers contain submodules that interact independently or redundantly with promoters. Conversely, indirect cooperativity is based on the equilibrium competition between nucleosomes and DNA-binding proteins, thereby not requiring protein-protein interactions. In the case of HIFmediated transcription, the binding of cooperating transcription factors has been demonstrated for several target genes. In particular, HIF-mediated expression of the erythropoietin gene requires an adjacent HNF4 binding site, and PAI-1 induction by hypoxia has been linked to cooperative promoter activation by CEBPa.
proteasomal degradation, heterodimerize with HIFb subunits and translocate to the cell nucleus, where they bind the RCGTG consensus sequence within regulatory regions of target genes, leading to their transcriptional activation in hypoxia. Mammals present three isoforms of HIFa that differ in their tissue distribution, HIF1a being the more ubiquitous and best characterized. A large number of studies focusing on single genes have identified individual HIF targets that, collectively, account for the functional responses to hypoxia, mainly metabolic adaptation and induction of angiogenesis. More recently, works employing HIF1a and HIF2a chromatin immunoprecipitation coupled to genomic microarrays or high-throughput sequencing have addressed the genome-wide identification of HIF binding locations, thereby improving the existing knowledge on the HIF-modulated transcriptome and largely confirming the RCGTG HIF binding consensus. Additionally, these studies have provided important insights into the global properties of HIF1 binding and transactivation. First, these works reported a significant association between the presence of a HIF binding site and hypoxic induction of the neighboring genes. The same trend was not found for genes repressed by hypoxia, Folinic acid calcium salt pentahydrate suggesting that hypoxia-mediated repression is largely indirect or HIF independent. Furthermore, they have clearly shown that only a small subset of about a hundred of all RCGTG-containing genes is robustly regulated by hypoxia. Hence, and in agreement with work on other transcription factors, HIFs bind a small proportion of potential binding sites, albeit the basis of their binding and target selectivity are incompletely understood. Understanding the mechanisms that explain HIFs transactivation selectivity is of paramount importance to expand our knowledge on transcriptional regulation and to improve the sensitivity and specificity of genome-wide efforts to characterize the HIF transcriptional response. DNA accessibility of transcription factor binding sites can clearly contribute to binding selectivity. For HIFs, recent evidence includes enhanced HIF1 and HIF2 binding to normoxic DNAse hypersensitivity sites and enrichment of HIF1 binding in the proximity of genes with a “permissive” transcriptional state in normoxia, as evidenced by significant basal expression. Additionally, DNA methylation has been also shown to modulate HIF1 binding, as originally demonstrated for the 39 enhancer of the Lomitapide Mesylate erythropoietin gene. A further mechanism that can impact target selectivity is direct or indirect cooperativity between transcription factors. Models of direct cooperativity have been mainly derived from developmental enhancers, and include the strict enhanceosome model, where cooperative occupancy occurs through extensive protein-protein interactions between TFs or common cofactors, and the more flexible billboard model, which suggests that enhancers contain submodules that interact independently or redundantly with promoters. Conversely, indirect cooperativity is based on the equilibrium competition between nucleosomes and DNA-binding proteins, thereby not requiring protein-protein interactions. In the case of HIFmediated transcription, the binding of cooperating transcription factors has been demonstrated for several target genes. In particular, HIF-mediated expression of the erythropoietin gene requires an adjacent HNF4 binding site, and PAI-1 induction by hypoxia has been linked to cooperative promoter activation by CEBPa.