The association between urine osmolarity and the risk of initiating dialysis, a stronger marker for end stage renal disease, has not been previously investigated. Our data thus adds to the existing evidence. The data is in line with Torres et al. suggesting a faster renal function decline in patients with higher urine osmolarity. Our cohort appeared to have the highest urine osmolarity with a median of 510 mosm/L versus a mean of 3686159 mosm/L, and a mean of 270 to 334 mosm/L. It is therefore conceivable that lowering urine osmolarity below a specific threshold might be harmful to the kidney as well. A recent study addressing the association between sodium excretion, an important part of total urine osmolarity, and end-stage renal disease in patients with type 1 diabetes mellitus showed an inverse association with end stage renal disease. Furthermore, a U-shaped curve was described for sodium excretion and mortality, such that subjects with the highest and the lowest sodium excretion had the highest mortality, partly supporting the hypothesis of low urine osmolarity being associated with harmful effects. Interestingly, in the univariate versus the Gomisin-D multivariable analysis the strong effect of impaired renal function seems to obscure the positive correlation of osmolarity with the incidence of ESRD. Adjusting for creatinine clearance reveals this relationship. We conclude that among patients with equal renal function those with higher osmolarity values are more likely to progress to ESRD than those with lower osmolarity values in 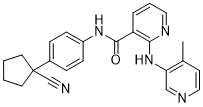 our cohort. As described in the methods, Danshensu estimated urine osmolarity was calculated from urine sodium, potassium and urea concentrations in 24 hour urine samples. These are the dominant solutes in the urine. Other solutes represent less than 10% of total urinary solutes. Thus, estimated urine osmolarity is often used as an approximation of true osmolarity. Regarding the difference between osmolality and osmolarity, it is negligible in our study because the density of urine is very close to that of pure water in the range of values considered here. Other authors have used estimated urine osmolarity in several studies. Several authors have studied urine volume and GFR decline. Opposed to the prevailing view that water is beneficial in CKD, Hebert et al. reported higher GFR decline in higher urine volume quartiles. However, multivariable adjustment seemed to diminish the association. In support of Hebert et al., Wang et al. found a weak, but significant association between higher urine volume and GFR decline. A large study by Clark et al. showed a relationship between higher urine volume and lower rate of eGFR decline, which stayed significant after multivariable adjustment. Another large study reported worse kidney function in individuals with a lower self-reported fluid intake;however, neither urine volume nor urine osmolarity were evaluated. It has been suggested that lower mean baseline GFR in cohorts of Hebert et al. and Wang et al., which is associated with alterations in water metabolism, might explain these paradoxical findings. With falling GFR, the ability to concentrate urine to osmolalities greater than that of plasma is progressively lost.
our cohort. As described in the methods, Danshensu estimated urine osmolarity was calculated from urine sodium, potassium and urea concentrations in 24 hour urine samples. These are the dominant solutes in the urine. Other solutes represent less than 10% of total urinary solutes. Thus, estimated urine osmolarity is often used as an approximation of true osmolarity. Regarding the difference between osmolality and osmolarity, it is negligible in our study because the density of urine is very close to that of pure water in the range of values considered here. Other authors have used estimated urine osmolarity in several studies. Several authors have studied urine volume and GFR decline. Opposed to the prevailing view that water is beneficial in CKD, Hebert et al. reported higher GFR decline in higher urine volume quartiles. However, multivariable adjustment seemed to diminish the association. In support of Hebert et al., Wang et al. found a weak, but significant association between higher urine volume and GFR decline. A large study by Clark et al. showed a relationship between higher urine volume and lower rate of eGFR decline, which stayed significant after multivariable adjustment. Another large study reported worse kidney function in individuals with a lower self-reported fluid intake;however, neither urine volume nor urine osmolarity were evaluated. It has been suggested that lower mean baseline GFR in cohorts of Hebert et al. and Wang et al., which is associated with alterations in water metabolism, might explain these paradoxical findings. With falling GFR, the ability to concentrate urine to osmolalities greater than that of plasma is progressively lost.
Month: April 2019
Comparable urinary solute dilution varies considerably between individuals
Interestingly, median 24-hour urine osmolality is greater than that of plasma in humans, suggesting continuous antidiuretic action, which has been associated with renal function decline. Consequentially, Wang et al. recently devised a quantitative method to determine the amount of water needed on a case-by-case basis to achieve a mean urine osmolality Ginsenoside-Ro equivalent to that of plasma. Relationships between urine osmolarity and GFR decline have been described in two studies with contrasting results. We were interested in studying urine volume and urine osmolarity in terms of harder endpoints in chronic kidney disease. Thus we set out to study these variables in terms of risk of initiating dialysis, with death as a competing event. To describe intra- versus inter-individual variance of urine osmolarity, we conducted a variance component analysis including all run-in urine osmolarity values that were available for each patient using a mixed model with patients as levels of a random factor. The outcome variable was time to dialysis, with death as the competing event. Patients who were alive without dialysis at the time of their last visit were censored. Absolute event rates were computed as the number of events divided by the total follow-up time for all patients. Observations with missing values were not used in the calculated models. We described the distribution of time to dialysis using cumulative incidence functions, and compared groups using Gray��s test. Due to the established relationship between baseline creatinine clearance and risk of initiating dialysis/ESRD, and the known progressive loss in urine concentration ability with decreasing renal function, it seemed important to introduce creatinine clearance 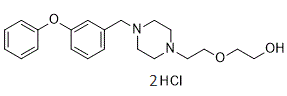 as an Gentiopicrin adjustment factor in all further analyses. We fi ed two multivariate proportional sub-distribution hazards models for competing risk data according to Fine and Gray in order to assess the effect of urine osmolarity or volume on the risk for initiating dialysis. In these models, we considered osmolarity or urine volume and included those variables that either proved significant in a multivariate model or changed the log hazard ratio of osmolarity or urine volume by more than 15% when those variables were excluded from the analyses. We assumed that any variable not selected would have no relevant impact on our conclusions. All variables listed in Table 1 were considered as potential confounders. Results from multivariate competing risk regression were described by means of sub-distribution hazard ratios and 95% confidence intervals, and by computing and visualising estimated cumulative incidence curves at specific covariate values. As urine osmolarity and creatinine clearance were log-base-2 transformed, their SHR correspond to each doubling of these variables. We checked for significant pairwise interactions of variables and for time-dependent effects by including interactions with follow-up time. Non-linear effects were assessed by the method of fractional polynomials. For sensitivity analysis, we also estimated a cause-specific Cox regression model. The R-software, version 2.12, was used for statistical analysis. Other models were calculated for follow-up urine osmolarities and did stay significant after further adjustments.
as an Gentiopicrin adjustment factor in all further analyses. We fi ed two multivariate proportional sub-distribution hazards models for competing risk data according to Fine and Gray in order to assess the effect of urine osmolarity or volume on the risk for initiating dialysis. In these models, we considered osmolarity or urine volume and included those variables that either proved significant in a multivariate model or changed the log hazard ratio of osmolarity or urine volume by more than 15% when those variables were excluded from the analyses. We assumed that any variable not selected would have no relevant impact on our conclusions. All variables listed in Table 1 were considered as potential confounders. Results from multivariate competing risk regression were described by means of sub-distribution hazard ratios and 95% confidence intervals, and by computing and visualising estimated cumulative incidence curves at specific covariate values. As urine osmolarity and creatinine clearance were log-base-2 transformed, their SHR correspond to each doubling of these variables. We checked for significant pairwise interactions of variables and for time-dependent effects by including interactions with follow-up time. Non-linear effects were assessed by the method of fractional polynomials. For sensitivity analysis, we also estimated a cause-specific Cox regression model. The R-software, version 2.12, was used for statistical analysis. Other models were calculated for follow-up urine osmolarities and did stay significant after further adjustments.
Large polyprotein that is subsequently processed by a combination of cellular and the viral NS2B/3 proteinase
To yield the three structural proteins capsid, pre-membrane and envelope and the nonstructural proteins, NS1, NS2A, NS2B, NS3, NS4A, 2K, NS4B and NS5. Replication of the DENV genome occurs in intimate association with perinuclear ER membranes which are modified to form characteristic structures during virus infection. High-throughput RNA interference studies have shown that DENV depends heavily on the cellular machinery for replication. However the mechanisms by which DENV interacts with cellular pathways and the viral and cellular proteins involved, largely remain to be determined. Comparative analysis of the gene expression profiles of a range of cell types infected with DENV in vitro and cells isolated from the blood of DENV infected individuals has identified a number of genes and cellular signaling pathways that are specifically dysregulated in DENV infection and may be Sipeimine involved in pathogenesis. In addition, high-throughput interaction studies have identified a number of interactions between DENV and cellular proteins that may play a role in replication or avoiding host defense mechanisms. By contrast to gene expression studies, the analysis of the host response to either DENV or flavivirus infection at the proteomic level is more limited. The standard approach of two-dimensional PAGE combined with the identification of specific proteins by mass spectrometry has been used to  detect proteins that are altered in amount in DENV infected mammalian cells, insect cells and in sera from DENV infected patients and resulted in the identification of a number of cellular proteins potentially relevant to pathogenesis. However this type of analysis is limited by the resolution and sensitivity of 2D-PAGE. In recent years, advances in the sensitivity of MS, coupled with high-throughput protein identification has made it feasible to quantify global Ursolic-acid changes in cellular protein levels in response to viral infection. The use of stable isotope labeling techniques to distinguish proteins derived from different cell populations, either by metabolic labeling of proteins or chemical modification of peptides, in combination with quantitative MS, provides the most sensitive means of accurately analyzing the proteome of a cell currently available. By combining differential labeling techniques with subcellular fractionation and quantitative MS, it is possible not only to measure changes in the amounts of proteins, but also to study changes in the cellular distribution of proteins, even if the total protein levels have not altered significantly. This approach is well suited to the comparative analysis of cell populations such as control and virus infected cells, but surprisingly there are very few reports of the application of these techniques to study viral pathogenesis and none for DENV. In this study we investigated the effects of DENV-2 infection on the host cell proteome of human A549 cells using SILAC in combination with high throughput liquid chromatography MS/MS. The mock and infected A549 cells were fractionated into nuclear and cytoplasmic extracts before analysis to identify proteins that redistribute between cellular compartments during infection and reduce the complexity of the analysis. We identified proteins that both increased and decreased in response to DENV2 infection.
detect proteins that are altered in amount in DENV infected mammalian cells, insect cells and in sera from DENV infected patients and resulted in the identification of a number of cellular proteins potentially relevant to pathogenesis. However this type of analysis is limited by the resolution and sensitivity of 2D-PAGE. In recent years, advances in the sensitivity of MS, coupled with high-throughput protein identification has made it feasible to quantify global Ursolic-acid changes in cellular protein levels in response to viral infection. The use of stable isotope labeling techniques to distinguish proteins derived from different cell populations, either by metabolic labeling of proteins or chemical modification of peptides, in combination with quantitative MS, provides the most sensitive means of accurately analyzing the proteome of a cell currently available. By combining differential labeling techniques with subcellular fractionation and quantitative MS, it is possible not only to measure changes in the amounts of proteins, but also to study changes in the cellular distribution of proteins, even if the total protein levels have not altered significantly. This approach is well suited to the comparative analysis of cell populations such as control and virus infected cells, but surprisingly there are very few reports of the application of these techniques to study viral pathogenesis and none for DENV. In this study we investigated the effects of DENV-2 infection on the host cell proteome of human A549 cells using SILAC in combination with high throughput liquid chromatography MS/MS. The mock and infected A549 cells were fractionated into nuclear and cytoplasmic extracts before analysis to identify proteins that redistribute between cellular compartments during infection and reduce the complexity of the analysis. We identified proteins that both increased and decreased in response to DENV2 infection.
Translation of the genome results in the production of a well established in 4-HNE
Formed by arachidonic acid or other unsaturated fa y acids Tetrahydroberberine following free Benzoylpaeoniflorin radicals a ack, can bind to proteins by Michael addition, with preferential amino acidic binding sites to cysteine, hystidine or lysine residues. Therefore, 4-HNE protein adducts could be considered a reliable OS markers and have a biological impact on protein function. In this regard, the amino acid sequence of the human beta-actin shows several potential 4-HNE binding sites on all the major conformational subdomains of the protein.  A total of 6 C, 8 H and 19 K residues are present in the sequence, and potential 4-HNE binding amino acid residues are present in all the relevant conformational subdomains of the protein, for a total of 2 C, 4 H and 7 K in the subdomain 1, a total of 1 H and 3 K in the subdomain 2, a total of 2 C, 2 H and 5 K in the subdomain 3, and a total of 3 C and 4 K in the subdomain 4. In particular, the subdomain 2 of the molecule is of critical importance for DNAase 1 binding and protein polymerization, while a very crucial C 374 residue and potential binding site for 4-HNE is present in the conformationally critical hydrophobic pocket of the molecule. Our findings indicate that the observed reduction in beta-actin band intensity is independent of the employed monoclonal antibodies and are compatible with a real decrease in the betaactin expression in the R patients erythrocytes membrane. Moreover, the reported evidence exhaustively addresses the question of whether the observed decrease in beta-actin protein expression might be related to oxidative PTMs potentially preventing antibody recognition. Taken together, our data indicate that major alterations exist in the beta-actin of R erythrocytes which results both from changes in expression and as a consequence of oxidative damage. To this regard, beta-actin is a well-known major target for OS processes, while erythrocytes represent ����cellular detectors���� revealing the co-existence of major cytoskeletal changes in this key genetic model for neurodevelopmental disorders. In particular, this work suggests a novel role for MeCP2 as a stabilizing protein in microtubule dynamics. To date, no definitive cure for R exists, although several approaches to a potential therapy have been either a empted or hypothesized, including activation of the silenced Mecp2 gene, gene therapy, modulation of some of the downstream effects from MeCP2-deficiency. Our study indicates that a proteomic approach in R is able to reveal additional downstream effects of the MECP2-deficiency, and therefore could identify potentially novel therapeutical targets for the disease. The four serotypes of dengue virus cause the most important arthropod-borne viral disease of humans. DENV infection results in a range of clinical outcomes ranging from the milder dengue fever to the potentially life threatening dengue haemorrhagic fever/dengue shock syndrome. A recent study estimates that up to 390 million people are infected with DENV annually, making dengue a serious global public-health problem. Despite much effort, there are neither vaccines nor antiviral therapies in clinical use to prevent or treat dengue, and our understanding of dengue pathogenesis is still limited. DENV is a member of the Flavivirus genus of the Flaviviridae family and has a RNA genome of,11 kb in size.
A total of 6 C, 8 H and 19 K residues are present in the sequence, and potential 4-HNE binding amino acid residues are present in all the relevant conformational subdomains of the protein, for a total of 2 C, 4 H and 7 K in the subdomain 1, a total of 1 H and 3 K in the subdomain 2, a total of 2 C, 2 H and 5 K in the subdomain 3, and a total of 3 C and 4 K in the subdomain 4. In particular, the subdomain 2 of the molecule is of critical importance for DNAase 1 binding and protein polymerization, while a very crucial C 374 residue and potential binding site for 4-HNE is present in the conformationally critical hydrophobic pocket of the molecule. Our findings indicate that the observed reduction in beta-actin band intensity is independent of the employed monoclonal antibodies and are compatible with a real decrease in the betaactin expression in the R patients erythrocytes membrane. Moreover, the reported evidence exhaustively addresses the question of whether the observed decrease in beta-actin protein expression might be related to oxidative PTMs potentially preventing antibody recognition. Taken together, our data indicate that major alterations exist in the beta-actin of R erythrocytes which results both from changes in expression and as a consequence of oxidative damage. To this regard, beta-actin is a well-known major target for OS processes, while erythrocytes represent ����cellular detectors���� revealing the co-existence of major cytoskeletal changes in this key genetic model for neurodevelopmental disorders. In particular, this work suggests a novel role for MeCP2 as a stabilizing protein in microtubule dynamics. To date, no definitive cure for R exists, although several approaches to a potential therapy have been either a empted or hypothesized, including activation of the silenced Mecp2 gene, gene therapy, modulation of some of the downstream effects from MeCP2-deficiency. Our study indicates that a proteomic approach in R is able to reveal additional downstream effects of the MECP2-deficiency, and therefore could identify potentially novel therapeutical targets for the disease. The four serotypes of dengue virus cause the most important arthropod-borne viral disease of humans. DENV infection results in a range of clinical outcomes ranging from the milder dengue fever to the potentially life threatening dengue haemorrhagic fever/dengue shock syndrome. A recent study estimates that up to 390 million people are infected with DENV annually, making dengue a serious global public-health problem. Despite much effort, there are neither vaccines nor antiviral therapies in clinical use to prevent or treat dengue, and our understanding of dengue pathogenesis is still limited. DENV is a member of the Flavivirus genus of the Flaviviridae family and has a RNA genome of,11 kb in size.
Many novel proteins not previously identified to be effected by DENV infection
Bioinformatic analysis was used to Praeruptorin-B identify a number of processes affected by DENV-2 infection. The results of the SILAC-MS analysis was validated for seven selected proteins by Western blo ing and immunofluorescence microscopy. This is the first report of the application of SILACMS  to study changes in the host cell proteome in response to DENV infection and demonstrates the power of this technique for identifying and quantifying changes in cellular protein amounts in response to DENV infection. In order to conduct the proteomic analysis, human lung carcinoma A549 cells were grown for eight cell doublings in either light or heavy Senegenin labeled media before being mock infected or infected with DENV-2 respectively. Although not believed to represent a target cell for DENV in vivo, A549 cells are highly permissive for DENV infection and have been used in previous studies examining the effect of DENV on the cellular innate immune response and the host cell transcriptome. At 28 hours p.i., a time point determined by growth curve analysis to lie in exponential phase of DENV replication, the cells were harvested and fractionated into nuclear and cytoplasmic extracts. At this time there was no obvious difference in the morphology of the mock and DENV infected cells, suggesting that there was li le cell death. IFA analysis of a sample of the mock and infected A549 cells revealed that 100% of the cells had been infected. The fractionation was done to reduce the overall complexity of the sample and to identify proteins that were altered in amount or redistributed between the cytoplasm and nucleus during DENV infection. DENV is known to replicate in tight association with perinuclear membranes therefore the cells were fractionated using a procedure that removed as much of the perinuclear membrane as possible without disrupting the nuclei. The presence of protein markers in the cellular fractions specific to the nucleus, cytoplasm and DENV infection were analyzed by Western blo ing to validate the infection and fractionation procedures. The analysis showed that whilst the nuclear and soluble cytoplasmic protein fractions were distinct, the lack of detergent in the lysis buffer led to the presence of some membraneous/ cytoskeletal proteins in the nuclear fractions. It appeared that the perinuclear membrane and the associated viral replication structures were not totally removed from the nuclei, as evidenced by a minor amount of the virus E protein, which is cytoplasmically localized, in the nuclear fraction. By contrast, the NS5 protein is known to be found in both the nucleus and cytoplasm of infected cells. Equal protein amounts from the cytoplasmic fractions of the mock and DENV-2 infected cells were pooled and the same procedure repeated for the nuclear fractions. The proteins in the cytoplasmic and nuclear fractions were separated by 1D SDSPAGE, subject to in-gel tryptic digestion and the peptides analyzed by quantitative LC-MS/MS to determine the relative amounts of proteins in the nuclear and cytoplasmic fractions from mock and DENV-2 infected cells. This procedure was done once and resulted in the identification of 4053 and 2881 cellular proteins in the cytoplasmic and nuclear fractions of which 3098 and 2115 respectively, were reliably quantified.
to study changes in the host cell proteome in response to DENV infection and demonstrates the power of this technique for identifying and quantifying changes in cellular protein amounts in response to DENV infection. In order to conduct the proteomic analysis, human lung carcinoma A549 cells were grown for eight cell doublings in either light or heavy Senegenin labeled media before being mock infected or infected with DENV-2 respectively. Although not believed to represent a target cell for DENV in vivo, A549 cells are highly permissive for DENV infection and have been used in previous studies examining the effect of DENV on the cellular innate immune response and the host cell transcriptome. At 28 hours p.i., a time point determined by growth curve analysis to lie in exponential phase of DENV replication, the cells were harvested and fractionated into nuclear and cytoplasmic extracts. At this time there was no obvious difference in the morphology of the mock and DENV infected cells, suggesting that there was li le cell death. IFA analysis of a sample of the mock and infected A549 cells revealed that 100% of the cells had been infected. The fractionation was done to reduce the overall complexity of the sample and to identify proteins that were altered in amount or redistributed between the cytoplasm and nucleus during DENV infection. DENV is known to replicate in tight association with perinuclear membranes therefore the cells were fractionated using a procedure that removed as much of the perinuclear membrane as possible without disrupting the nuclei. The presence of protein markers in the cellular fractions specific to the nucleus, cytoplasm and DENV infection were analyzed by Western blo ing to validate the infection and fractionation procedures. The analysis showed that whilst the nuclear and soluble cytoplasmic protein fractions were distinct, the lack of detergent in the lysis buffer led to the presence of some membraneous/ cytoskeletal proteins in the nuclear fractions. It appeared that the perinuclear membrane and the associated viral replication structures were not totally removed from the nuclei, as evidenced by a minor amount of the virus E protein, which is cytoplasmically localized, in the nuclear fraction. By contrast, the NS5 protein is known to be found in both the nucleus and cytoplasm of infected cells. Equal protein amounts from the cytoplasmic fractions of the mock and DENV-2 infected cells were pooled and the same procedure repeated for the nuclear fractions. The proteins in the cytoplasmic and nuclear fractions were separated by 1D SDSPAGE, subject to in-gel tryptic digestion and the peptides analyzed by quantitative LC-MS/MS to determine the relative amounts of proteins in the nuclear and cytoplasmic fractions from mock and DENV-2 infected cells. This procedure was done once and resulted in the identification of 4053 and 2881 cellular proteins in the cytoplasmic and nuclear fractions of which 3098 and 2115 respectively, were reliably quantified.